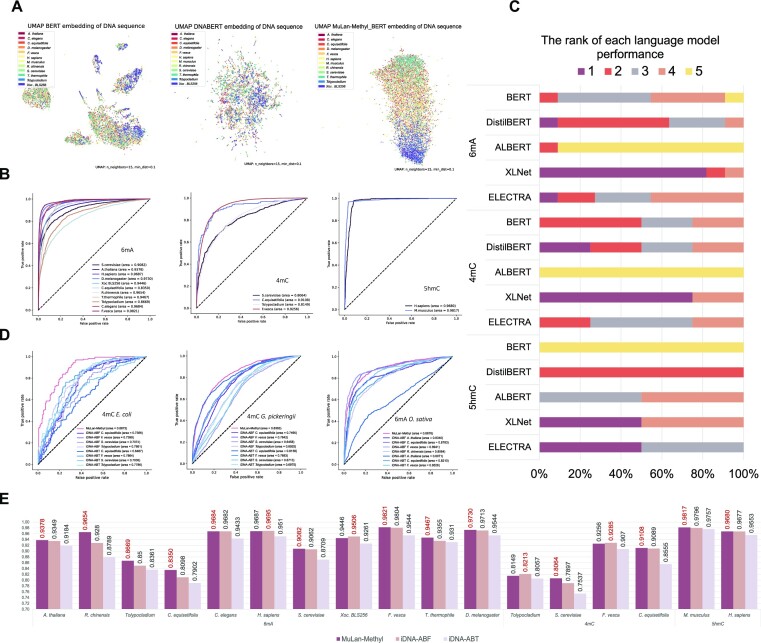
Figure 2:
Model analysis and performance comparison of MuLan-Methyl. (A) UMAP clustering of sample representations encoded by different pretrained models: BERT, DNABERT, and MuLan-Methyl_BERT (from left to right). Samples are colored by taxonomic lineage. (B) For MuLan-Methyl predictions of the 3 methylation site types, 6mA, 4mC, and 5hmC, we present ROC curves for each of the 12 taxonomic types in the dataset. The AUC values are shown in brackets. (C) For each of the 3 methylation site types and each of the 5 language models, BERT, DistilBERT, ALBERT, XLNet, and ELECTRA, we show the ranking of models over all taxonomic lineages in terms of AUC scores. Moreover, the frequency with which each fine-tuned model appeared is indicated as the width of the corresponding block. (D) Comparison of MuLan-Methyl against 2 published methods, iDNA-ABF and iDNA-ABT, on an additional dataset that only contains taxonomic lineages that were not used to train the methods. From left to right, we show the ROCs obtained for the prediction of 4mC sites in E. coli, 4mC sites in G. pickeringii data, and 6mA sites in O. sativa L. data, respectively. (E) Comparison of MuLan-Methyl against iDNA-ABF and iDNA-ABT, on the iDMA-MS independent test set. We display the AUC scores for all 3 methods, for each of the 3 methylation site types and each of the 12 taxonomic lineages.