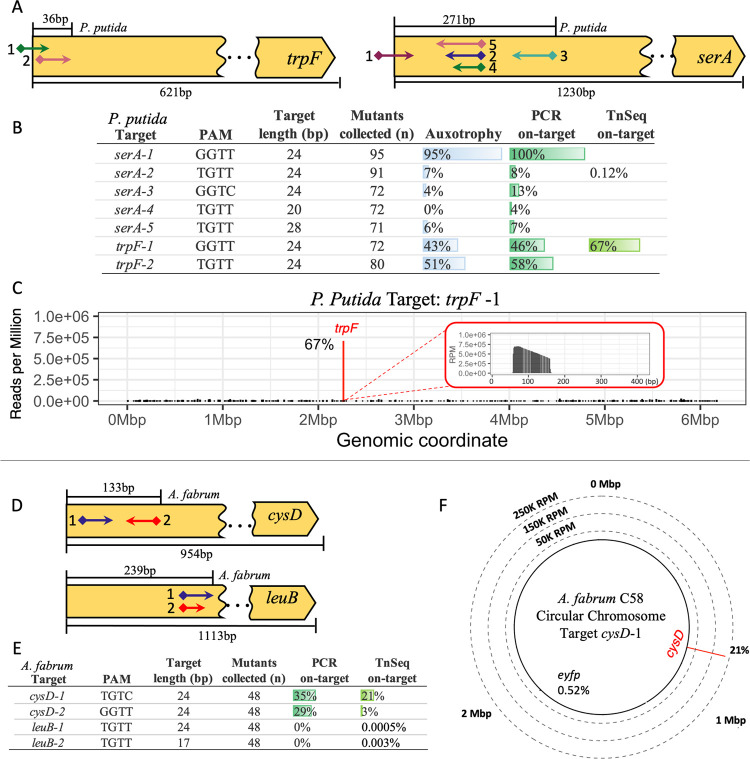
Figure 4.
Gene disruption in P. putida and A. fabrum. (A) Map of auxotrophic sgRNA targets across P. putida amino acid biosynthesis genes trpF and serA. SgRNA targets (arrows) with the PAM site (diamonds) and insertion direction (arrow point). SgRNA targeting range (above gene) and total gene length (below gene). (B) Relative frequency table of auxotrophy complementation (n = mutants collected), genotype (n = 24), and Tn-Seq results for P. putida targets. (C) Tn-Seq results for P. putida target trpF-1 with on-target reads (red bar) and frequency (%) across the whole genome (bin size = 1 Kbp). Zoomed-in view of transposon insertion of trpF-1 start at target site PAM (0bp) (bin size = 1 bp). (D) Map of sgRNA targets across A. fabrum amino acid biosynthesis genes cysD and leuB. (E) Relative frequency table of genotype (n = 48) and Tn-Seq results for A. fabrum targets. (F) Tn-Seq results for A. fabrum target cysD-1 with on-target reads (red bar) and frequency (%) across the whole genome (bin size = 1 Kbp). Dashed rings represent the Y-axis in reads per million (RPM) with Mbp (Megabase pair) denoting the chromosome coordinates.