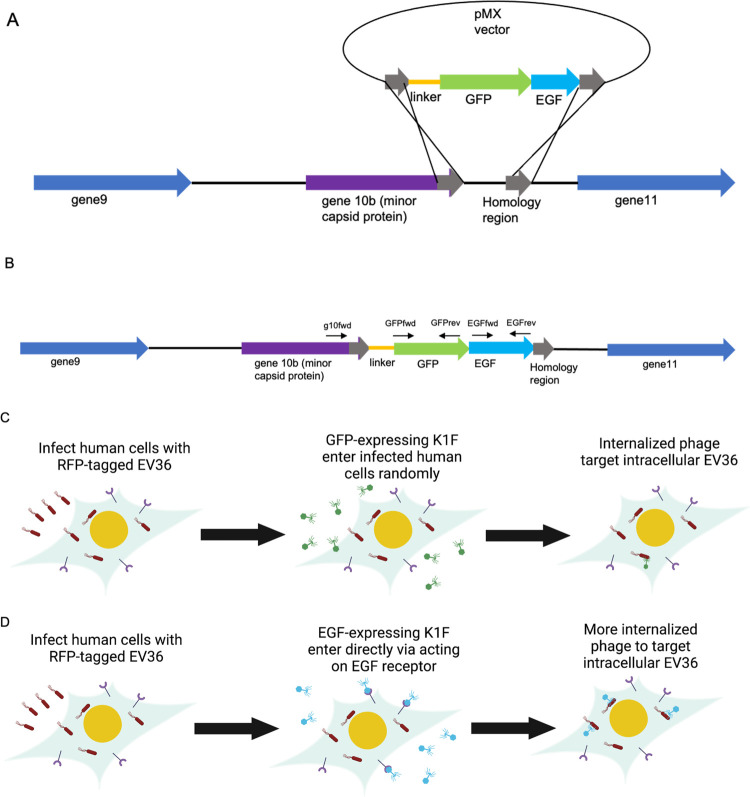
Figure 1.
Schematic of the genome engineering of bacteriophage K1F to express GFP and EGF. (A) Engineering method to generate the synthetic phage. This was performed via an in vivo homologous recombination method with plasmid pMX:GFP-EGF serving as the donor DNA. Phage infection initiates a double crossover homologous recombination event between the homologous regions (gray), leading to the integration of the GFP-EGF protein fusion at the C-terminus of g10b (purple). (B) Schematic of the final construct generated as a single translational fusion. Primers used to probe for the presence and correct orientation of genes are denoted above the respective genes shown as black arrows. (C) Schematic detailing the in vitro system. Human cell lines are infected with EV36 and treated with K1F-GFP, which enter human cells randomly via phagocytosis, targeting intracellular EV36. Addition of EGF to K1F allows directed entry via receptor-mediated endocytosis (D), allowing higher quantities of phage to be internalized to clear intracellular bacteria within infected cells.