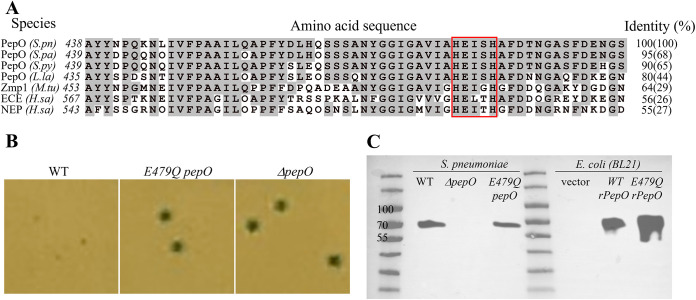
FIG 5.
The glutamic acid residue in the conserved “HExxH” catalytic motif is critical for PepO’s quorum quenching activity. (A) Sequence and alignment of S. pneumoniae PepO and its homologs surrounding the HExxH motif (red box). Gray-highlighted residues are identical to those of the S. pneumoniae PepO. Sequences and NCBI accession numbers are as follows: S.pn, Streptococcus pneumoniae PepO, WP_054394657.1; S.pa, Streptococcus parasanguinis PepO, MBF1717056.1; S.py, Streptococcus pyogenes PepO, WP_136303158.1; L.la, Lactococcus lactis PepO, WP_143457511.1; M.tu, Mycobacterium tuberculosis Zmp1, KBK61972.1; H.sa, Homo sapiens ECE isoform 1, BAG59124.1; H.sa, Homo sapiens NEP, AAI43466.1. Sequence identities are compared for the 58-amino-acid stretch surrounding the HExxH motif (and full-length protein). (B) Blue/white colony phenotypes of pneumococcal strains (Pshp1518-lacZ) expressing wild-type PepO, E479Q PepO, or ΔpepO in CDM (glucose) agar. (C) Western blotting of wild-type and mutant PepO expressed in S. pneumoniae or E. coli BL21. Samples are prepared from whole-cell lysates of Pshp1518-lacZ S. pneumoniae strains containing the indicated genotype or E. coli BL21 strains. For blotting, the membrane was incubated overnight at 4°C with a rabbit anti-PepO antibody (1:1,000) raised against pneumococcal PepO (43) followed by a 30-min incubation with a goat anti-rabbit horseradish peroxidase (HRP) secondary antibody (1:10,000). The sizes of native PepO and rPepO are 71.9 kDa and 74.5 kDa, respectively.