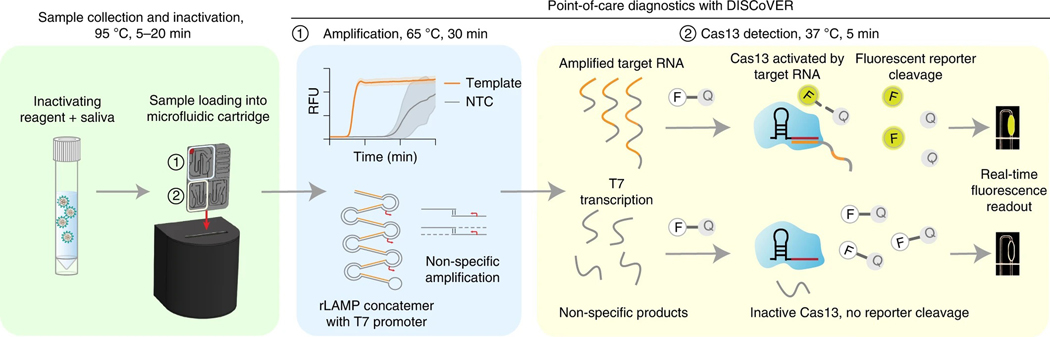
Fig. 1 |. DISCoVER microfluidic system for rapid and automated molecular diagnostics.
Patient samples, such as saliva, are collected and heat-inactivated in direct lysis buffer, followed by loading onto a single-use, gravity-driven microfluidic cartridge. The inactivation step ranges from 5 to 20 min depending on institutional regulations37. The cartridge is then inserted into a companion instrument that automatically runs the DISCoVER assay in a closed system to minimize reaction contamination. In step 1, an initial rLAMP reaction employs two mechanisms for amplification of target nucleic acids. RFU, relative fluorescence units. Cas13 enzymes are programmed with a guide RNA to specifically recognize the desired RNA molecules over non-specifically amplified products. Subsequent activation of Cas13 ribonuclease activity, in step 2, results in cleavage of reporter molecules for saturated signals within 5 min of CRISPR detection. Including actuation time of the device, time to readout in a finalized system is 48 min. In the left half of the cartridge, guide RNAs targeting SARS-CoV-2 enable rapid and selective detection of attomolar concentrations of virus. The mirrored half of the cartridge is used for an internal process control, enabling a negative test result by ensuring the presence of adequate patient samples. By exploiting template switching and CRISPR programmability, the point-of-care DISCoVER system can contribute to increased surveillance of diverse pathogens.