Figure 2. Influence of riboswitches on ribonuclease attack.
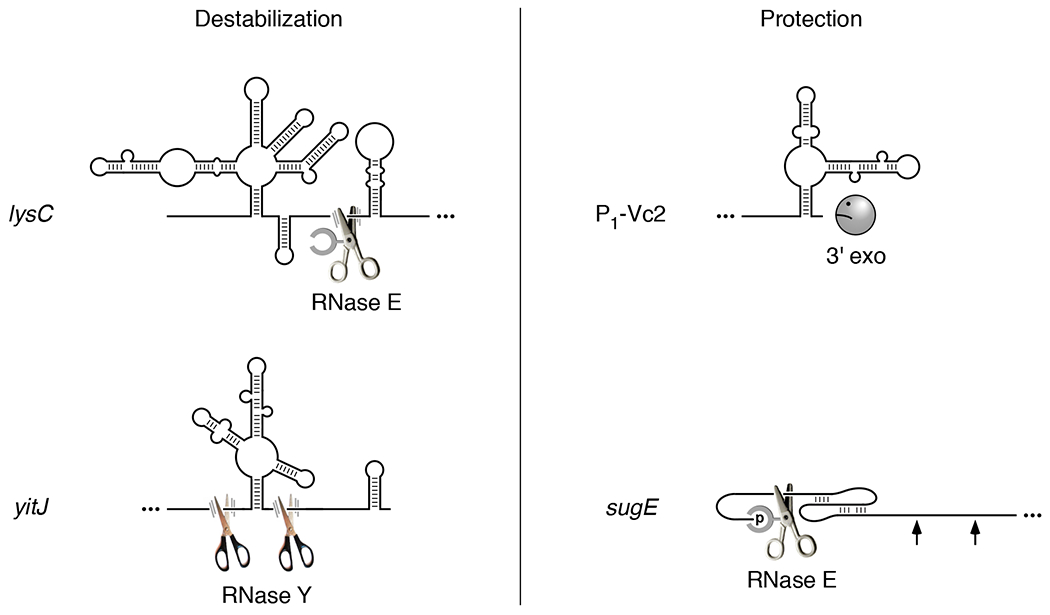
(Left) Ligand-induced RNA destabilization. Ligand binding to the E. coli lysC or B. subtilis yitJ riboswitch results in an RNA conformational change that exposes previously inaccessible cleavage sites for RNase E or RNase Y, respectively, two endonucleases that cut at specific sites in single-stranded regions of RNA. Cleavage at only one of the two adjacent RNase E cleavage sites in the lysC 5′ UTR is shown. (Right) Ligand-induced RNA protection. (Upper right) The conformational change caused by ligand binding to the V. cholerae Vc2 riboswitch is thought to protect P1-Vc2 RNA from 3′-exonucleolytic attack by sequestering its 3′ end in base pairing. (Lower right) Ligand-induced formation of a coaxially base-paired pseudoknot by the guanidine-III riboswitch of L. pneumophila sugE mRNA blocks access to distal sites by RNase E, which binds the monophosphorylated 5′ end of the transcript and scans linearly from there in search of cleavage sites. All four riboswitches are depicted in the base-paired conformation they adopt upon ligand binding (McDaniel et al., 2003, Kulshina et al., 2009, Smith et al., 2009, Caron et al., 2012, Sherlock & Breaker, 2017). Scissors, endonuclease. Frowning Pac-Man, 3′ exonuclease deterred by a base-paired 3′ end. Arrow, inaccessible cleavage site. Ellipsis, RNA segment not shown. Parallel lines representing motion blur identify endonucleases able to cut lysC and yitJ RNA at the indicated sites. The absence of such lines in the case of sugE signifies a scanning endonuclease (RNase E) that is stalled at a site it cannot cut.
