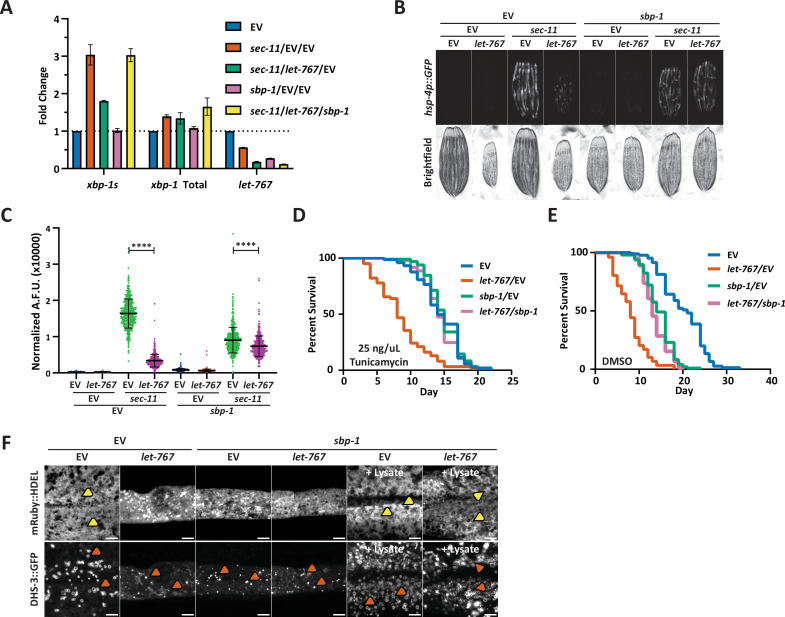
Figure 3. Reduced global lipid synthesis rescues the UPRER suppression caused by let-767 RNAi.
(A) Quantitative RT-PCR transcript levels of xbp-1 total, xbp-1s, and let-767 from day 1 adult N2 animals grown from L1 on EV, sec-11 and EV, let-767 and sec-11 RNAi combined with either EV or sbp-1 RNAi in a 1:1:1 ratio. Fold-change compared to EV treated N2 animals. Lines represent standard deviation across three biological replicates, each averaged from two technical replicates. (B) Fluorescent micrographs of day 1 adult transgenic animals expressing hsp-4p::GFP grown on EV, let-767, sec-11, and/or sbp-1 RNAi mixed in a 1:1:1 ratio to assay effects on the UPRER induction. (C) Quantification of (D) normalized to size using a BioSorter. Lines represent mean and standard deviation. n=426. Mann-Whitney test p-value ****<0.0001. Representative data shown is one of three biological replicates. (D–E) Concurrent survival assays of N2 animals transferred to ER stress conditions of 25 ng/uL Tunicamycin (D) or control DMSO (E) conditions at day 1 of adulthood. Animals continuously grown on EV or let-767 RNAi combined in 1:1 ratio with EV or sbp-1 RNAi from L1 synchronization. (F) Representative fluorescent micrograph projections of day 1 adult transgenic animal expressing ER lumen-localized mRuby::HDEL or LD-localized dhs-3::GFP, grown on EV or let-767 RNAi mixed in 1:1 ratio with sbp-1 RNAi with or without N2 lysate supplementation to assay ER and LD quality. Yellow arrowheads point to wide ER structures. Orange arrowheads point to lipid droplets. Scale bar, 5 μm. Images for organelle markers individually contrasted for clarity.