To the Editor—We read with interest the article by Rauseo et al [1] regarding the derivation and internal validation of a clinical predictive model for fluconazole resistance in patients hospitalized with candidemia. Their model consisted of 5 clinical parameters associated with fluconazole-resistant candidemia: older age, hematopoietic stem cell transplant (HSCT), myelodysplastic syndrome (MDS), azole exposure, and recent bacteremia. The authors propose this model may identify patients who could safely receive first-line or early step-down fluconazole therapy without awaiting susceptibility testing. External validation of this model in an international cohort is desirable before its clinical implementation, especially considering previous difficulties validating similar risk models in invasive candidiasis [2]. We aimed to assess the generalizability of Rauseo et al's [1] model to an Australian context and explore clinical factors predictive of fluconazole resistance in an Australian setting.
We performed a retrospective cohort study at Austin Hospital, a 671-bed tertiary care academic hospital in Melbourne, Australia, which undertakes HSCT and renal and liver transplantation. All adult patients hospitalized and aged ≥18 years with Candida spp isolated from ≥1 blood culture between January 1, 2017 and January 1, 2023 were included. This study was approved by our institution's Human Research Ethics Committee (HREC/92327). We adhered to the same study protocol used by Rauseo et al [1], including the same data collection methods and endpoints as published. We also collected data on variables not included in the original study (Supplementary Data). We fitted the same model as described by Rauseo et al [1], and we evaluated its discrimination and calibration. In addition, an alternative risk prediction model was developed using backward stepwise logistic regression (including variables with P < .20 on univariable logistic regression) in 1000 bootstrapped samples, and the final model included variables present in ≥65% samples. Model performance was evaluated by C-statistic, Hosmer-Lemeshow test, and calibration plot. The final score was calculated using logit coefficients and diagnostic performance at each cutoff is presented. Data were analyzed in STATA (StataCorp, College Station, TX).
Candidemia occurred in 111 patients. Twenty-five patients (22.5%) had fluconazole-resistant isolates as defined by Clinical and Laboratory Standards Institute performance standards for antifungal testing [3]. Median age of all subjects was 64 years (interquartile range, 54–75) and 53 (47%) were female. The fluconazole-resistant group had higher rates of systemic azole exposure within the last year (10 [40%], 10 [12%], P = .002) and neutropenia ≤1000 cells/µL within the last 30 days (9 [36%], 13 [15%], P = .025) (Supplementary Table 1).
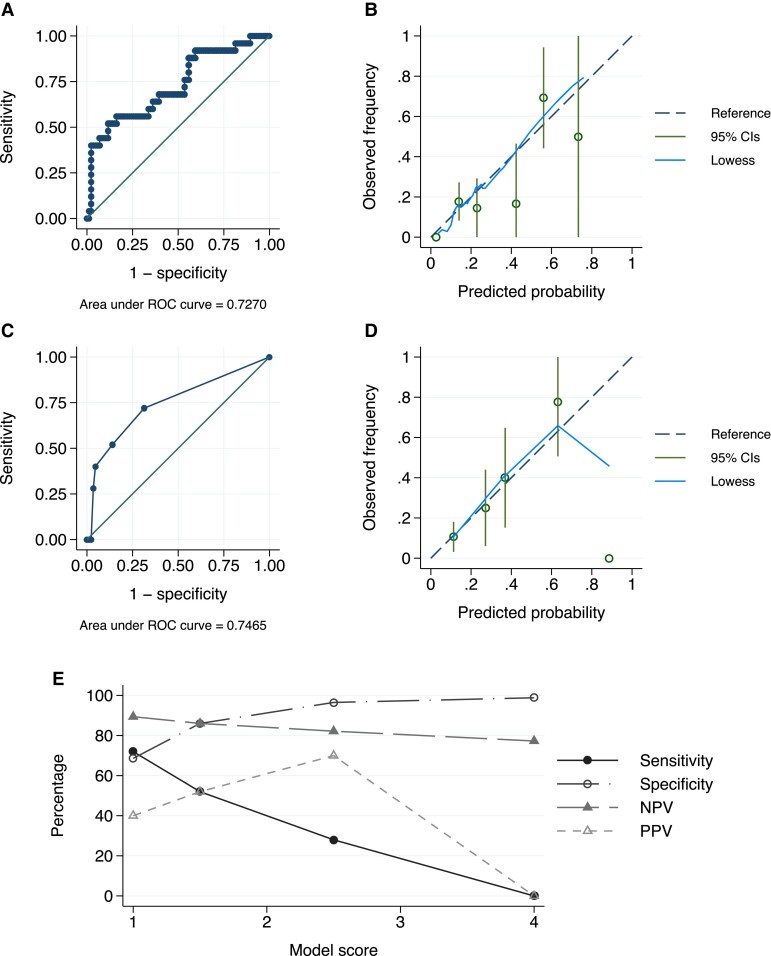
When the model proposed by Rauseo et al [1] was applied to our Australian cohort, the C-statistic was 0.727 (compared to 0.788) (Figure 1AandB).
Figure 1.
(A) Receiver operating characteristic (ROC) curve for validation of the model by Rauseo et al [1], regression predicting candidemia; (B) calibration plot for the model by Rauseo et al [1]; (C) ROC curve for our clinical predictive model; (D) calibration plot for our model; (E) final score and associated model performance where 1.5 points is assigned for intensive care unit admission, 1 point for Charlson comorbidity index score 3–4, and 1.5 points for azole use. Highest sensitivity achieved with 1 point (72%, specificity 69%, positive predictive value [PPV] 40%, negative predictive value [NPV] 89%) and highest PPV achieved with 2.5 points (70%, sensitivity 28%, specificity 97%, NPV 82%). CI, confidence interval.
The alternative model consisted of 3 variables significantly associated with prediction of fluconazole-resistant candidemia: systemic azole exposure within the last year (odds ratio [OR] = 4.60, 95% confidence interval [CI] = 1.54–13.75, P = .006, assigned 1.5 points to the score), Charlson comorbidity index (CCI) score 3–4 (OR = 2.93, 95% CI = 1.07–8.00, P = .037, 1 point), and transfer to the intensive care unit (ICU) within 48 hours of blood culture collection, which was selected a priori as a marker of critical illness (OR = 4.61, 95% CI = .74–28.8, P = .102, 1.5 points). The C-statistic was 0.747, the Hosmer-Lemmeshow test was 0.380, and calibration was acceptable (Figure 1CandD). Diagnostic performance is shown in Figure 1E.
Azole exposure was the only similarity shared with the Rauseo et al [1] model. This is a well described risk factor supported by in vitro, retrospective, and prospective cohort studies around the world [4–6]. In contrast, hematological factors (HSCT, MDS) were not significant predictors. This may be explained by different prophylaxis strategies, with posaconazole preferred in Australian settings and fluconazole used more commonly in some North American centers [7]. We also did not find older age or recent bacteremia to be predictive of fluconazole resistance. Novel predictors in our study included multiple medical comorbidities (CCI score 3–4) and critical illness (ICU transfer within 48 hours of blood culture collection). Despite variables differing between studies, all variables may be considered surrogate measures of an individual's functional reserve or disease severity, and this association between fluconazole resistance and frail/sick patients remains consistent between studies and across the wider literature [8, 9]. The exact mechanism is unclear and may be due to unmeasured confounding, which cannot be taken into account.
Unfortunately, our findings are not consistent with those presented by Rauseo et al [1], emphasizing the need for local epidemiological data before the adoption of existing international risk predictive models. The lessons learned from other candida risk-prediction and colonization indices may again be at play—highlighting that local epidemiology, antifungal consumption, and host factors continue to limit the holy grail of a one-size-fits all clinical decision rule approach to candidemia and predicting resistant phenotypes.
Supplementary Material
Contributor Information
Brennan Collis, Department of Infectious Diseases, Austin Health, Heidelberg, Victoria, Australia.
Sara Vogrin, Department of Medicine, University of Melbourne, Parkville, Victoria, Australia.
Jason A Trubiano, Department of Infectious Diseases, Austin Health, Heidelberg, Victoria, Australia; Department of Medicine, University of Melbourne, Parkville, Victoria, Australia; National Centre for Infections in Cancer, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia; Department of Infectious Diseases, The Peter Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, Australia.
Gemma Reynolds, Department of Infectious Diseases, Austin Health, Heidelberg, Victoria, Australia; Department of Medicine, University of Melbourne, Parkville, Victoria, Australia; National Centre for Infections in Cancer, Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Rauseo AM, Olsen MA, Stwalley D, et al. . Creation and internal validation of a clinical predictive model for fluconazole resistance in patients with Candida bloodstream infection. Open Forum Infect Dis 2022; 9:ofac447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Playford EG, Lipman J, Kabir M, et al. . Assessment of clinical risk predictive rules for invasive candidiasis in a prospective multicentre cohort of ICU patients. Intensive Care Med 2009; 35:2141–5. [DOI] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute . Performance standards for antifungal susceptibility testing of yeasts. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 4. Andes D, Forrest A, Lepak A, Nett J, Marchillo K, Lincoln L. Impact of antimicrobial dosing regimen on evolution of drug resistance in vivo: fluconazole and Candida albicans. Antimicrob Agents Chemother 2006; 50:2374–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slavin MA, Sorrell TC, Marriott D, et al. . Candidaemia in adult cancer patients: risks for fluconazole-resistant isolates and death. J Antimicrob Chemother 2010; 65:1042–51. [DOI] [PubMed] [Google Scholar]
- 6. Cuervo G, Puig-Asensio M, Garcia-Vidal C, et al. . A simple prediction score for estimating the risk of candidaemia caused by fluconazole non-susceptible strains. Clin Microbiol Infect 2015; 21:684.e1–9. [DOI] [PubMed] [Google Scholar]
- 7. Freifeld AG, Bow EJ, Sepkowitz KA, et al. . Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 2011; 52:e56–93. [DOI] [PubMed] [Google Scholar]
- 8. Oxman DA, Chow JK, Frendl G, et al. . Candidaemia associated with decreased in vitro fluconazole susceptibility: is Candida speciation predictive of the susceptibility pattern? J Antimicrob Chemother 2010; 65:1460–5. [DOI] [PubMed] [Google Scholar]
- 9. Ostrosky-Zeichner L, Harrington R, Azie N, et al. . A risk score for fluconazole failure among patients with candidemia. Antimicrob Agents Chemother 2017; 61:e02091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.