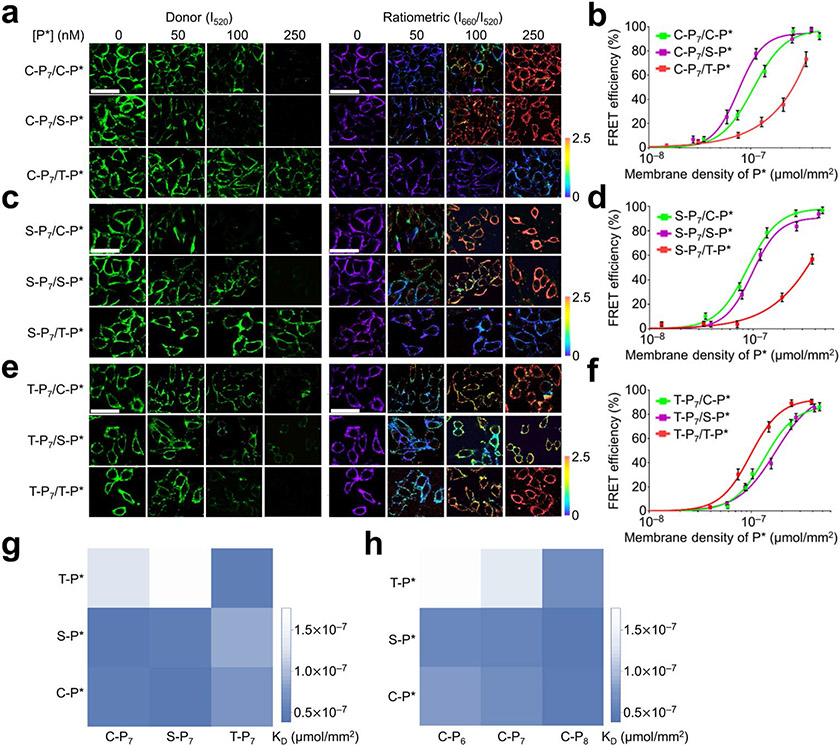
Figure 2.
Characterization of DNA Zippers on MCF-7 cell membranes. (a) 100 nM of C-P7 probe, (c) 90 nMof S-P7 probe, and (e) 90 nMof T-P7 probe was incubated with 0–250 nM of C-P*, C-P*, or C-P* probe on the MCF-7 cells for 20 min at 25°C. Samples were then imaged at the same temperature via excitation via a 488 nm laser and the fluorescence emission of Atto488 (I520) and Cy5 (I660) was collected at ~520 and ~660 nm, respectively. Pseudo-color I660/I520 ratiometric images were also shown. Scale bar, 20 μm. (b, d, f) The FRET efficiency was calculated based on the changes in the donor fluorescence intensity. Shown are the mean and SEM values from at least 20 individual cells in each P* concentration, (g) Apparent membrane dissociation constant, KD, for each pair of lipid-DNA conjugates was shown. (h) The effect of DNA duplex length on the membrane DNA Zipper binding efficiency. C-P7, S-P7, and T-P7 represent cholesterol-, sphingomyelin-, and tocopherol-modified DNA donor probes. C-P6, C-P7, and C-P8 represent cholesterol-DNA donor probe with 6, 7, or 8-nucleotide-long hybridization region with the acceptor probe modified with cholesterol (C-P*), sphingomyelin (S-P*), or tocopherol (T-P*).