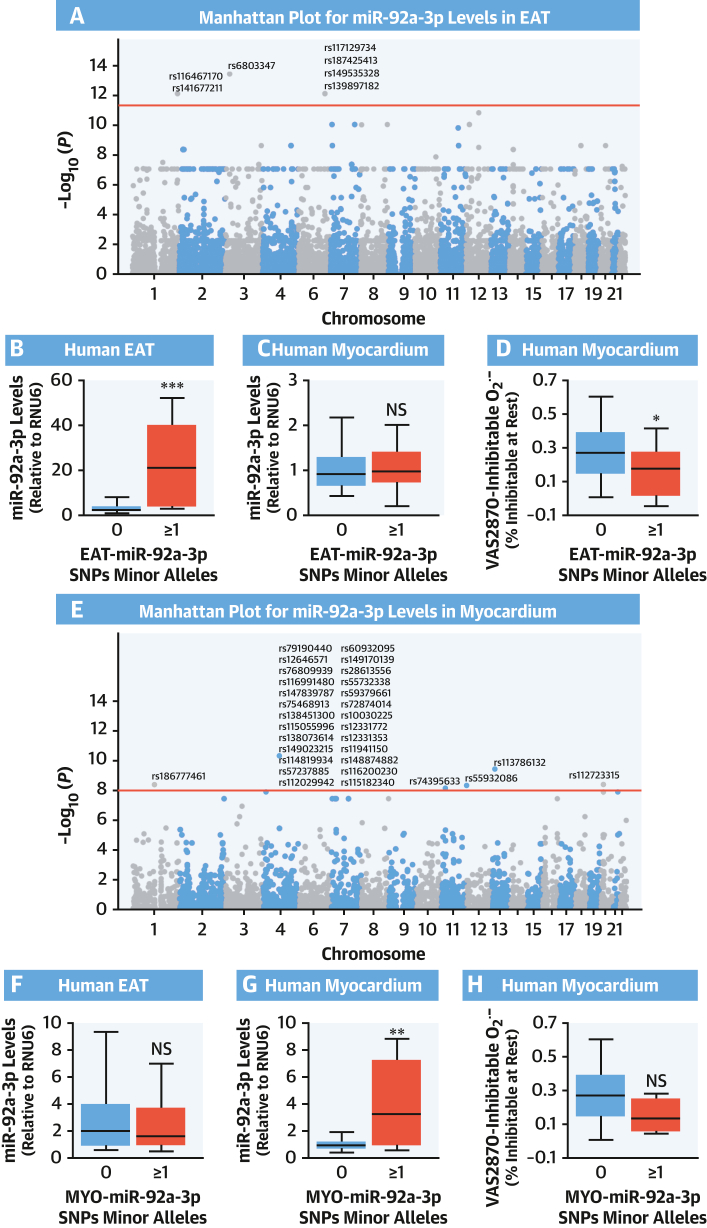
Figure 2.
SNPs Affecting EAT miR-92a-3p and Myocardial Redox State
Manhattan plots representing single-nucleotide polymorphisms (SNPs) associated with miR-92a-3p levels in EAT (A) and myocardium (E). Seven SNPs were significantly associated with high miR-92a-3p levels in EAT (EAT-miR-92a-3p) and 31 with high levels of miR-92a-3p in the myocardium (MYO-miR-92a-3p). The presence of any SNP from EAT-miR-92a-3p was associated with higher miR-92a-3p levels only in EAT, not in the myocardium (B, n = 149; C, n = 265), whereas the presence of any MYO-miR-92a-3p SNP was associated with higher miR-92a-3p levels only in the myocardium, not in EAT (G, n = 265; F, n = 149). The presence of any EAT-miR-92a-3p SNP led to a statistically significant reduction of myocardial superoxide production (D and H, n = 196). In B to D and F to H, data are presented as median (25th-75th percentile). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by the Mann-Whitney U test. NS = not significant; other abbreviations as in Figure 1.