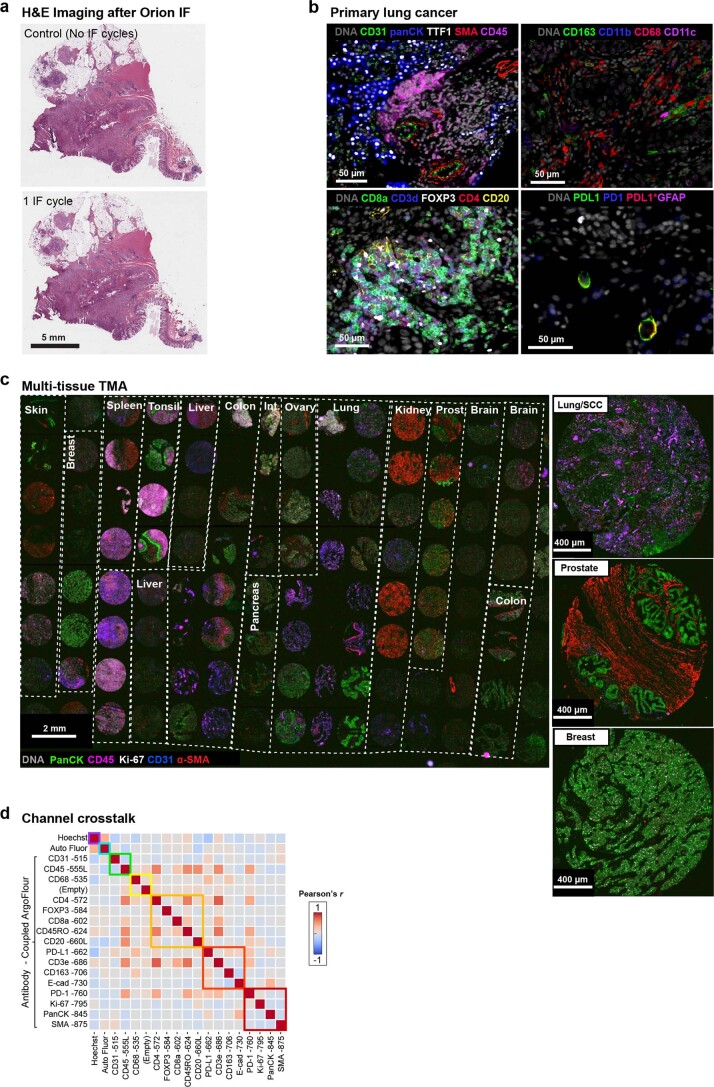
Extended Data Fig. 2. Orion imaging of different cancer types (colorectal and lung) and assessment of channel crosstalk.
a. Images of H&E-stained sections of colorectal cancer performed before IF imaging (no IF cycles) and after one cycle of IF imaging (1 IF cycle) showing excellent preservation of staining intensity and morphology. Scalebars 5 mm. Images from one sample. b, Representative images of 20-plex Orion panel from a primary lung adenocarcinoma sample. Note: two PD-L1 antibodies were used, PD-L1 (green) is E1L3N clone from Cell Signaling and PD-L1*(red) is EPR19759 from Abcam. Scalebars 50 µm. Images from one sample. c, 16-plex (18 channel) Orion image from a tissue microarray (TMA) containing normal and diseased human tissues including inflammatory and neoplastic diseases (Examples highlighted are lung squamous cell carcinoma (SCC), prostate adenocarcinoma, and breast ductal carcinoma); DNA, pan-cytokeratin, Ki-67, α-SMA, CD45 and CD31 are displayed. Scalebars 2 mm and 400 µm, as indicated. Images from one TMA containing 123 patient samples. d, Validation of minimal channel crosstalk in 18-plex tonsil image after spectral extraction. Pearson’s correlation coefficients between all channel pairs were calculated using the paired pixel intensities. Square boxes with colored borders denote excitation lasers. High correlation coefficients were only found in channel pairs that contains target markers that are in close proximity. Data was derived from a selected frame of (N =1) image.