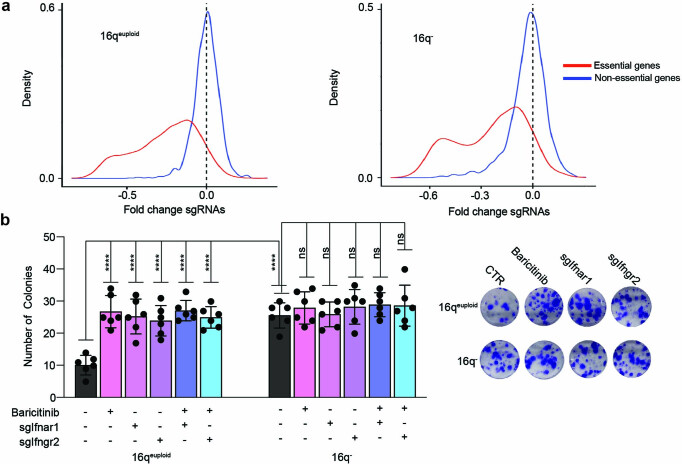
Extended Data Fig. 7. IFNRs are tumor suppressive in a cell autonomous manner.
a) Genome-Wide CRISPR Screen quality control via fold change separation curves generated using a previously curated list of known essential and non-essential genes. Comparison of the foldchange of guide level abundance at 20 doublings to the reference timepoint reveals significant drop-out in the essential genes and minimal drop out in the non-essential population, indicating no change from the reference population. b) Colony forming unity assay showing number of colonies after Ifnra1 or Ifngr2 knockouts with or without Baricitinib treatment compared to parental untreated cells (left panel) and representative images of the experiment (right panel), data are presented as mean values +/- SD (N = 6 tumors per each condition), p values = 4.73*10-5, 0.0012, 5.17*10-4, 1.25*10-5, 1.47*10-4. **** P < 0.0001 by two-way Anova with Tukey’s multiple comparison (b).