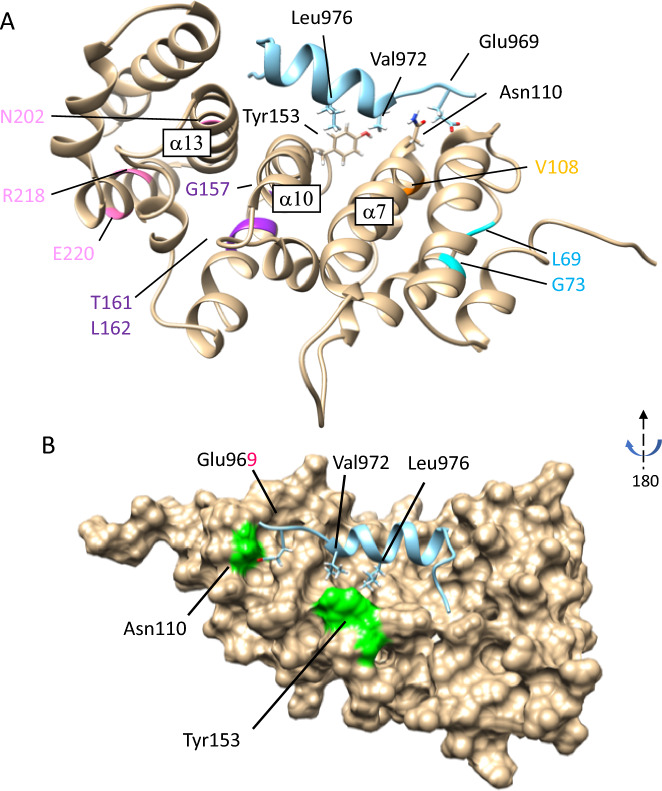
Figure 3.
Structural modeling of human INF2. A 3D structure of human INF2 modeled on the mDia1 DID-DAD complex crystal structure (PDB 2F31)16 using the Rosetta program. (A) Locations for ten pathogenic variants in the DID are illustrated as ribbon diagrams; Blue: L69P, G73D (G73V), Orange: V108D, Magenta: G157R, T161N, L162P, Pink: N202S, R218W, E220K. Based on this model, the α helix of the N-terminal DAD (turquois) is predicted to contact the core pocket of DID. Side chains of selected residues involved in the DID/DAD interactions are shown in stick form: N110 and Y153 in DID, E969, V972, and L976 in the DAD. The central α helices, α7, α10, α13, from the 2nd, third, fourth armadillo repeats respectively, create the DID-binding pockets. (B) Core DID-DAD interaction. The DID is shown as a surface representation rotated horizontally by 180˚. The residues E969, V972, L976 in the N-terminal DAD are predicted to make direct contact with the surface-exposed partners N110, Y153 in the hydrophobic binding pocket of DID.