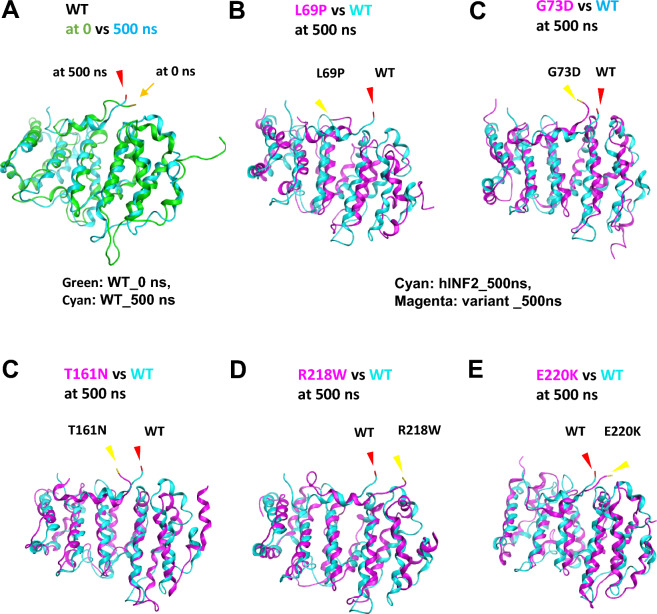
Figure 6.
Comparison of the residue mobility at the DID-DAD interface between wild-type INF2 and five pathogenic variants. Molecular dynamics of human INF2 structures are simulated using GROMACS program, assuming that the protein is solvated in water with salt. Images of dynamic structures are captured with VMD (http://www.ks.uiuc.edu/Research/vmd/) and are compared between wild-type INF2 (WT) with pathogenic variants. The DAD residue Glu968, which is involved in the DID-DAD interaction, is indicated as an index tracking marker (arrow, at t = 0 ns; arrowheads, at t = 500 ns). (A) Mobility of Glu968 residue in wild-type INF2. Superimposed images at 0 ns (green) and 500 ns (cyan) are shown. The position of Glu968 at 500 ns (arrowhead, red) deviates from that of the reference at time 0 ns (arrow yellow), indicating that the Glu 968 residue of wild-type INF2 is mobile during the 500 ns-MD simulation. (B–E) Mobility of Glu968 residue in pathogenic INF2 variants. Dynamic structures of wild-type (cyan) and INF2 variants (L69P, G73D, T161N, R218W, E220K, magenta) are simulated at 500 ns and the superimposed images are shown. For all five pathogenic INF2 variants, the positions of Glu968 at 500 ns (arrowhead, yellow) are displaced from the reference location of WT (arrowhead, red), indicating that the pathogenic variants have the altered mobility at the DID-DAD interface compared to the WT.