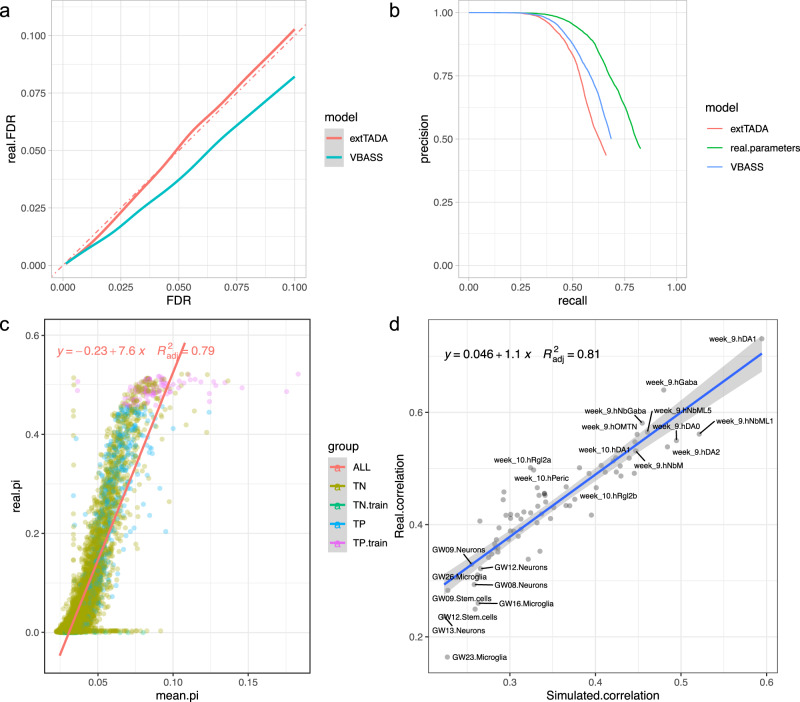
Fig. 3. Performance of single cell version VBASS on simulation data.
a Plot of true false discovery rate (real.FDR, y axis) at different FDR cutoff (x axis) estimated by extTADA and VBASS. Genes in the training data were removed. b Comparison of precision recall for extTADA and VBASS, only shown for the part with FDR ≤0.5. Genes in the training data were removed. c Scatter plot of disease risk prior () that we assigned in simulation (y-axis) and informed by VBASS (x-axis). Genes were colored by labels and whether used in semi-supervised training, where TN and TP correspond to true negative and true positive, respectively. d Comparison of correlation between real disease risk prior and cell type expression (y-axis) versus correlation between VBASS informed prior and cell type expression (x-axis). Each dot represents a cell type. Gray shading showed confidence interval of linear regression estimated by stat_smooth function in R ggplot2 package.