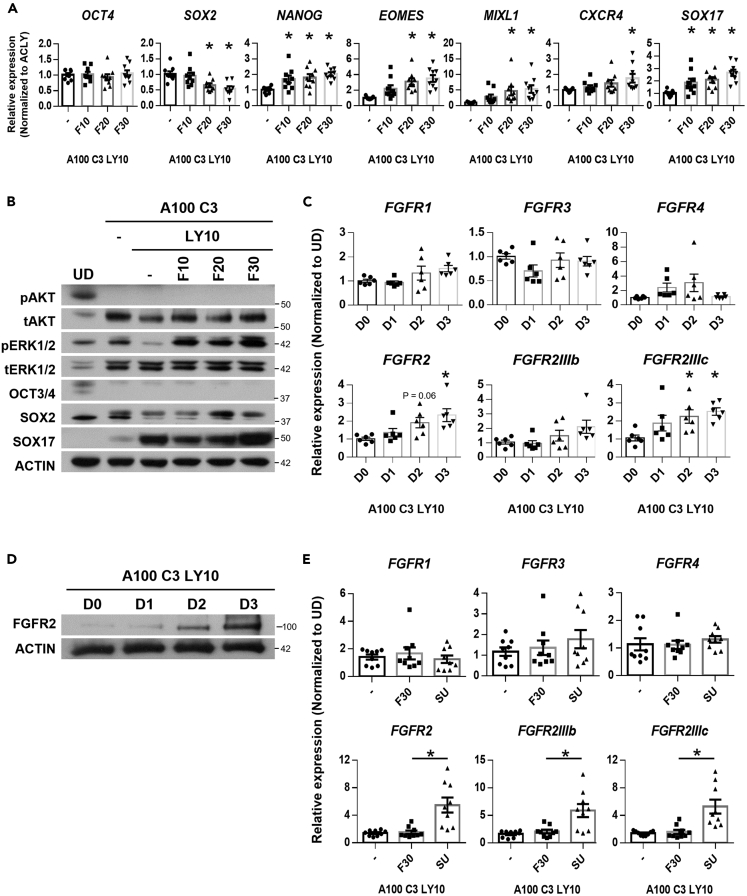
Figure 1.
FGF2-FGFR signaling is involved in DE formation
(A) Expression of pluripotency markers OCT4, SOX2, NANOG, mesendoderm markers EOMES, MIXL1 and DE markers CXCR4 and SOX17 on FGF2 supplementation at 10, 20 or 30 ng/mL (F10 – 30) in hPSCs differentiated into D3 DE using 100 ng/mL Activin A (A100), 3 μM CHIR99021 (C3) and 10 μM LY294002 (LY10). Gene expression is normalized to ACLY control. N=3 independent experiments were performed.
(B) Western blot assessment of proteins involved in D3 DE differentiation on FGF2 supplementation (F10 – 30). N=5 independent experiments were performed.
(C) Expression of FGF receptors (FGFR1, FGFR3, FGFR4, FGFR2, FGFR2IIIb, and FGFR2IIIc) across three days of DE differentiation. Gene expression is normalized to undifferentiated hPSCs (UD).
(D) Western blot assessment of FGFR2 protein expression across three days of DE differentiation. N=3 independent experiments were performed.
(E) Expression of FGF receptors (FGFR1, FGFR3, FGFR4, FGFR2, FGFR2IIIb, and FGFR2IIIc) on 30 ng/mL FGF2 (F30) or 10 μM SU5402 (SU) supplementation in hPSCs differentiated into D3 DE. Gene expression is normalized to UD. N=3 independent experiments were performed. All error bars in Figure 1 indicate standard error of mean of biological replicates. One-way ANOVA and Dunnett’s post-hoc tests were used for statistical analyses. Asterisk (∗) indicates p < 0.05 against each control group respectively. H9 hESCs were used for all experiments in Figure 1. “See also Figure S1.”