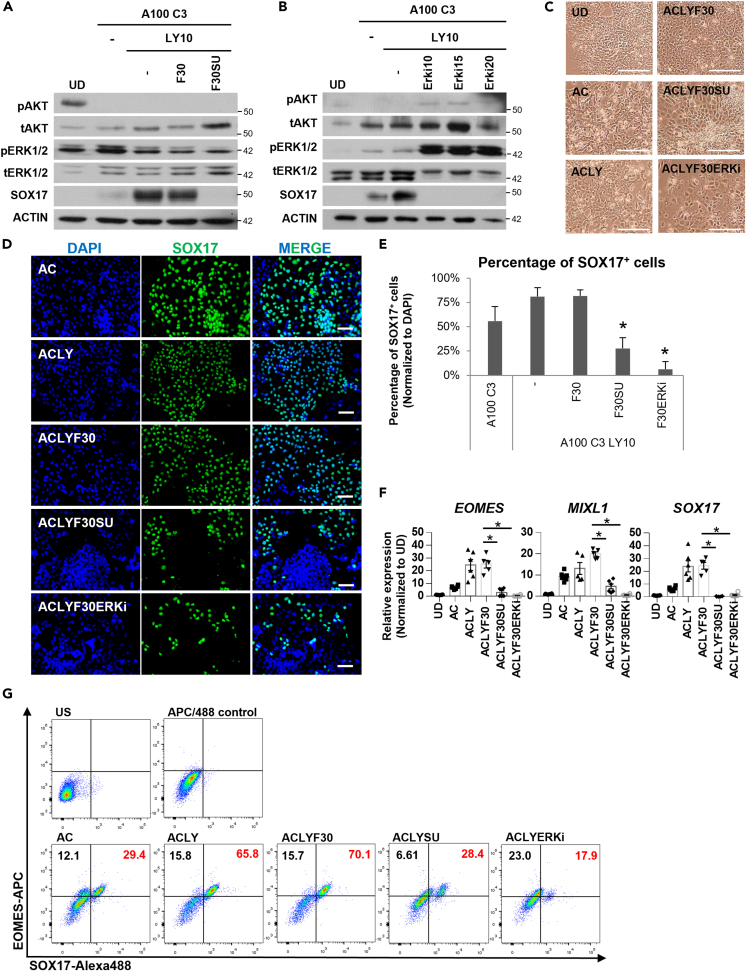
Figure 2.
FGF drives DE formation through FGFR and ERK1/2 signaling
Western blot assessment of proteins involved in D3 DE differentiation on (A) F30 or F30SU supplementation or (B) ERK inhibitor (ERKi) supplementation (10–20 μM). N=3 independent experiments were performed.
(C) Morphology of cells at the end of D3 DE differentiation. Scale bar: 100 μm.
(D) Immunostaining for SOX17 DE protein (green) in hPSCs differentiated into D3 DE with AC, ACLY, ACLYF30, ACLYF30SU or ACLYF30ERKi. DAPI (blue) stains for nuclei. Scale bar: 100 μm.
(E) The percentage of SOX17+ DE cells (green) in hPSCs differentiated into D3 DE with AC, ACLY, ACLYF30, ACLYF30SU or ACLYF30ERKi. Cell count from at least three images was tabulated for each treatment condition. Asterisk (∗) indicates p < 0.05 against ACLY. N=3 independent experiments were performed.
(F) Expression of mesendoderm markers EOMES, MIXL1 and DE marker SOX17 on F30, F30SU or F30ERK inhibitor supplementation in hPSCs differentiated into D3 DE. Gene expression is normalized to UD. N=3 independent experiments were performed.
(G) Flow cytometry assessment of cells co-expressing EOMES and SOX17 on day 3 under AC, ACLY, ACLYF30, ACLYSU or ACLYERKi differentiation condition. One-way ANOVA and Dunnett’s post-hoc tests were used for statistical analyses. All error bars indicate standard error of mean of biological replicates. Asterisk (∗) indicates p < 0.05 against F30. H9 hESCs were used for all experiments in Figure 2. “See also Figures S2 and S3.”.