Highlights
-
•
OS microenvironment displays heterogeneity in cellular composition and myeloid cells were the most commonly represented cell type.
-
•
Oosteoclasts in the OS microenvironment could be differentiated from myeloid cells.
-
•
Co-expression of receptors and ligands for intercellular communication is associated with patient prognosis.
-
•
Intercellular communication differs in tumor microenvironment of OS patients received chemotherapy or not.
Keywords: Osteosarcoma, Microenvironment, Intercellular communication, Single-cell sequencing
Abstract
Osteosarcoma (OS) is the most common primary bone cancer in children and young adults, patient survival rates have not improved in recent decades. To further understand the interrelationship between different cell types in the tumor microenvironment of osteosarcoma, we comprehensively analyzed single-cell sequencing data from six patients with untreated osteosarcoma. Nine major cell types were identified from a total of 46,046 cells based on unbiased clustering of gene expression profiles and canonical markers. Osteosarcoma from different patients display heterogeneity in cellular composition. Myeloid cells were the most commonly represented cell type, followed by osteoblastic and TILs. Copy number variation (CNV) results identified amplifications and deletions in malignant osteoblastic cells and fibroblasts. Trajectory analysis based on RNA velocity showed that osteoclasts in the OS microenvironment could be differentiated from myeloid cells. Furthermore, we explored the intercellular communications in OS microenvironment and identified multiple ligand-receptor pairs between myeloid cells, osteoblastic cells and their cells, including 21 ligand-receptor pair genes that significantly associated with survival outcomes. Importantly, we found chemotherapy may have an effect on cellular communication in the OS microenvironment by analyzing single-cell sequencing data from seven primary osteosarcoma patients who received chemotherapy. We believe these observations will improve our understanding of potential mechanisms of microenvironment contributions to OS progression and help identify potential targets for new treatment development in the future.
1. Introduction
Osteosarcoma (OS) is the most common primary bone cancer in children and young adults 10 to 30 years of age, with a peak incidence during the adolescent growth spurt[1], [2]. OS is characterized by high malignancy, strong invasiveness and extremely high mortality rate. Patient survival rates have not been significantly improved in recent decades, surgical resection combined with adjuvant chemotherapy is still the widely used treatment regimen[1], [3], [4]. Immunotherapeutic strategies have been proposed with encouraging results in a variety of tumors in recent years[5], [6], [7]. Liposomal muramyl tripeptide (MTP-PE) has been approved for the treatment of non-metastatic resectable osteosarcoma. MTP-PE exerts its antitumor effects by activating monocytes/macrophages in patients, supporting the promise of immunomodulation in OS therapy[8]. The development of new immunotherapeutic strategies will require a deeper understanding of the complex immune microenvironment of osteosarcoma, which is influenced not only by immune cells and vascular cells, but also by the complex network of cellular and signaling pathways associated with bone formation and remodeling [9], [10], [11]. However, the dynamics of the different cell types, intercellular communication and their impact on OS growth and patient prognosis have not been elucidated.
Single-cell genomics provides powerful new tools for exploring tumor genetic and functional heterogeneity, predicting evolutionary lineages and identifying rare cell subpopulations [12], [13]. Single-cell RNA sequencing (scRNA-seq) technology provides strong support for the analysis of human tumor heterogeneity and different subpopulations, and has proven to be key to elucidating tumor development and progression mechanisms [14], [15], [16]. With the maturation of single cell isolation techniques in the tumor microenvironment, the availability of high quality scRNA-seq data, and the proposed computational models for bioinformatic analysis [17], [18], a deeper exploration of the complexity of OS tumor microenvironment and intercellular communication has become possible.
In the present study, we comprehensively analyzed scRNA-seq data from six patients with untreated osteosarcoma and identified nine cell types based on unbiased clustering of gene expression profiles and canonical markers. We identified malignant osteoblastic cells and fibroblasts by copy number variation (CNV) and trajectory analysis. Based on receptor-ligand interactions, detailed analysis of intercellular communication associated with osteoblastic cells and myeloid cell was performed. Our data revealed a number of immune function and osteogenesis related factors that play important roles in intercellular communication in the OS microenvironment, correlating with patient prognosis. We also analyzed scRNA-seq data from seven OS patients received chemotherapy and compared the similarities and differences of their microenvironment with treatment naive patients. By decoding the potential origin of some cell types and their communication, we provide molecular information important to advance our understanding of OS microenvironment.
2. Methods
2.1. scRNA-seq data preprocessing
The raw scRNA sequencing data of human treatment naïve osteosarcoma was downloaded from GEO (PRJNA681896, n = 6). The raw data were processed by CellRanger 3.0 (10X Genomics) with default parameters and aligned to GRCh38 to generate UMI matrix for the downstream analysis. Expression matrix was further analyzed by Seurat 4.2 package for quality control and downstream analysis following the standard workflow with default settings. Low-quality cells that had less than 200 genes per cell and less than 3 cells per gene were discarded. Then the cells with feature counts over 50,000 or less than 200, and percentage of mitochondrial genes less than 0.25 were filtered out.
2.2. Dimension reduction, clustering, and identification of DEGs
The top 2000 most highly variable features that exhibit high cell-to-cell variation from each sample were selected for data integration. Canonical correlation analysis was applied to remove the batch effect for data integration. Next, the top 15 principal components of the integrated data were selected for principal component analysis, UMAP analysis, and graph-based clustering (resolution = 0.5) to identify distinct subpopulations. DEGs were identified by the ‘FindAllMarkers’ function in Seurat (min.pct = 0.25, thresh.use = 0.25). ‘Clusterprofile’ package was used for pathway enrichment analysis.
2.3. RNA velocity
RNA velocity was introduced to calculate the spliced and unspliced RNAs to indicate the transcriptional kinetic activity. A loom file with counts divided into spliced/unspliced/ambiguous of each gene in each cell was generated by velocyto.py on the BAM file from the CellRanger analysis. Seurat object were retained for downstream analysis. Then RNA velocity was estimated by velocyto.R with default settings. The velocity fields were projected on to the UMAP embedding from the Seurat analysis.
2.4. Infer copy number Variation(CNV) and cell–cell communications
InferCNV package (v1.3.3) was applied to infer CNV from scRNA-seq data, with cutoff = 0.1. We then combined the CNV classification with the UMAP-based and the gene-set based classifications to define the malignant cells and normal immune cells. The marker genes for macrophages, T cells, and oligodendrocytes were obtained from Neftel et al. (GSE131928). To investigate the molecular interaction networks among each cell subtype, CellPhoneDB (v2.1.7) algorithm was used to infer intercellular communication in the tumor environment. Single-cell sequencing data from seven primary OS patients (GSE152048) who had received chemotherapy were used to compare the differences in cellular communication in the microenvironment of OS patients with or without chemotherapy.
2.5. Survival analysis in OS patients
We collected HTSeq-Count and clinical data for 88 OS patients from GDC data portal (https://portal.gdc.cancer.gov/). The ligand and receptor pairs with significance in cell to cell crosstalk were filtered in survival analysis using ggforest function in ‘survminer’ R package. We have generated the forest plot for the top 10 ligand to receptor interactions in crosstalk between different cell types.
3. Results
3.1. Clustering analysis of osteosarcoma single cell sequencing data
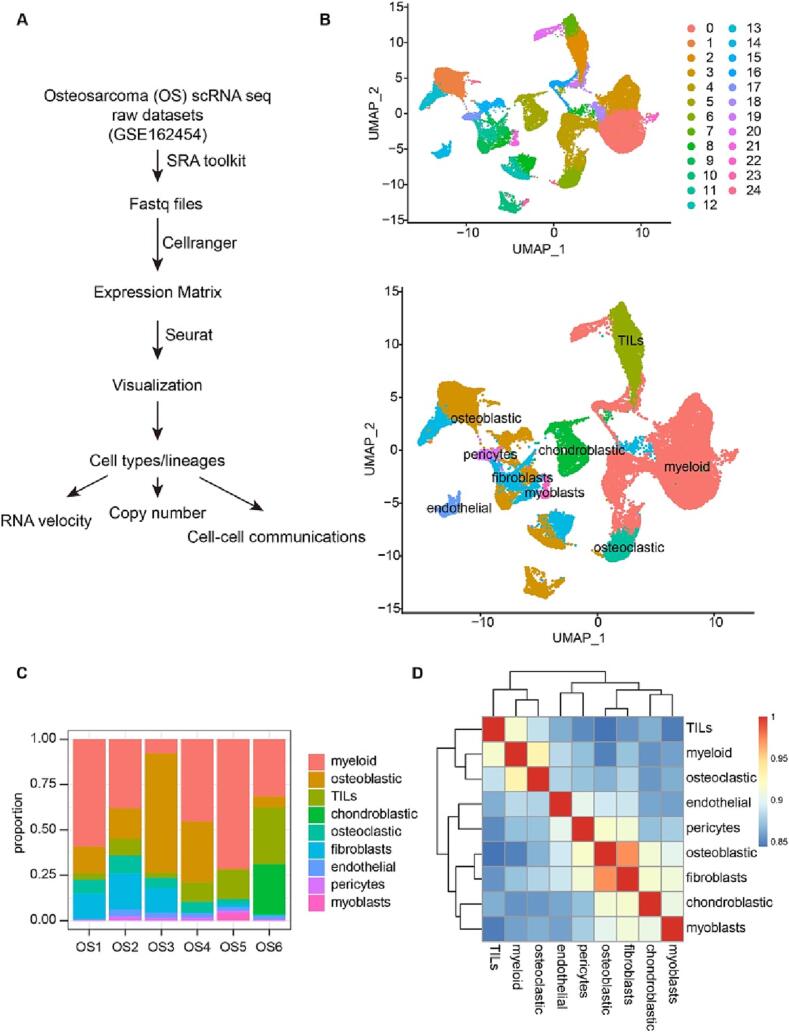
To explore the RNA velocity and cell–cell communication in OS tumor microenvironment, we analyzed single-cell sequencing data from six patients (P1-P6) with untreated osteosarcoma GSE162454 [1]. (Fig. 1A). The histological diagnosis of all patients was classical osteosarcoma. Five of them were adolescent patients aged 13–20 years and one was a 45-year-old middle-aged female. These fresh tumor tissues were taken from approximately 2 cm from the tumor margin and were digested and single-cell prepared after removal of fat, visible blood vessels and surrounding necrotic areas. Cell clustering and classification were based on based on t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) analyses according to their gene profiles and canonical markers [19]. In particular, marker genes used in this study were as follows: (1) the osteoblastic cells characterized with high COL1A1, CDH11 and RUNX2 expression; (2) the chondroblasts highly expressing ACAN, COL2A1 and SOX9; (3) the osteoclastic cells specifically expressing CTSK and MMP9; (4) the myeloid cells highly expressing CD74, CD14 and FCGR3A; (6) the fibroblasts defined by COL1A1, LUM and DCN; (5) the TILs highly expressing T and NK cell markers CD3D, IL7R and NKG7; (7) the pericytes highly expressing α-SMA and RGS5; (8) the mesenchymal stem cells (MSCs) expressing CXCL12, SFRP2 and MME; (9) the myoblasts characterized with high MYL1 and MYLPF; and (10) the endothelial cells identified by PECAM1 and VWF.
Fig. 1.
Identification of different cell types from OS single cell sequencing data. A. Graphical view of the study roadmap. B. The t-SNE plot of 24 clusters and 9 identified cell types in OS lesions. C. Proportion of different cell types in tumor tissues across the patients. D. The correlation heatmap illustrates the relationship between the different cell types infiltrated in OS.
Pooled analysis of all samples yielded a total of 46,046 cells organized into 24 clusters, which contained myeloid cells (19622), osteoblastic (9809), TILs (5943), chondroblastic (2447), osteoclastic (2337), fibroblasts (3907), myoblasts (4 0 3), endothelial (9 7 9) and pericytes (5 9 9) (Fig. 1B). The number of MSC cells was too small to be clustered out. Myeloid cells were the most commonly represented cell type (53%), followed by osteoblastic (21%) and TILs (14%). The proportions of cell populations were similar in all patients, except for P3 (Fig. 1C). The highest cell proportion in P3 was osteoblastic cells, while the percentage of myeloid cells was less than 10% (Fig. 1C). This may be related to the heterogeneity of the tumor and the individual variability of patients.
We then performed a correlation analysis of these cell populations in OS based on gene expression, and we found that there is a strong correlation at the population level between osteoblastic and fibroblast (Fig. 1D). Then, we analyzed the characteristic genes of different cell types. Highly expressed signature genes in each cell type were used to confirm cell types (Fig. 1B). Specifically, TILs expressed T cell and NK cell markers CD3D, IL7R, NKG7, endothelial expressed PECAM1 and VMF, while pericytes highly expressed ACTA2, RGS5 and MMP9 (Fig. 2). The top 10 differentially expressed genes (DEGs) for the 9 types of cells were shown in supplementary table 1.
Fig. 2.
The violin plots showing the normalized expression levels of signature genes across the identified cell types.
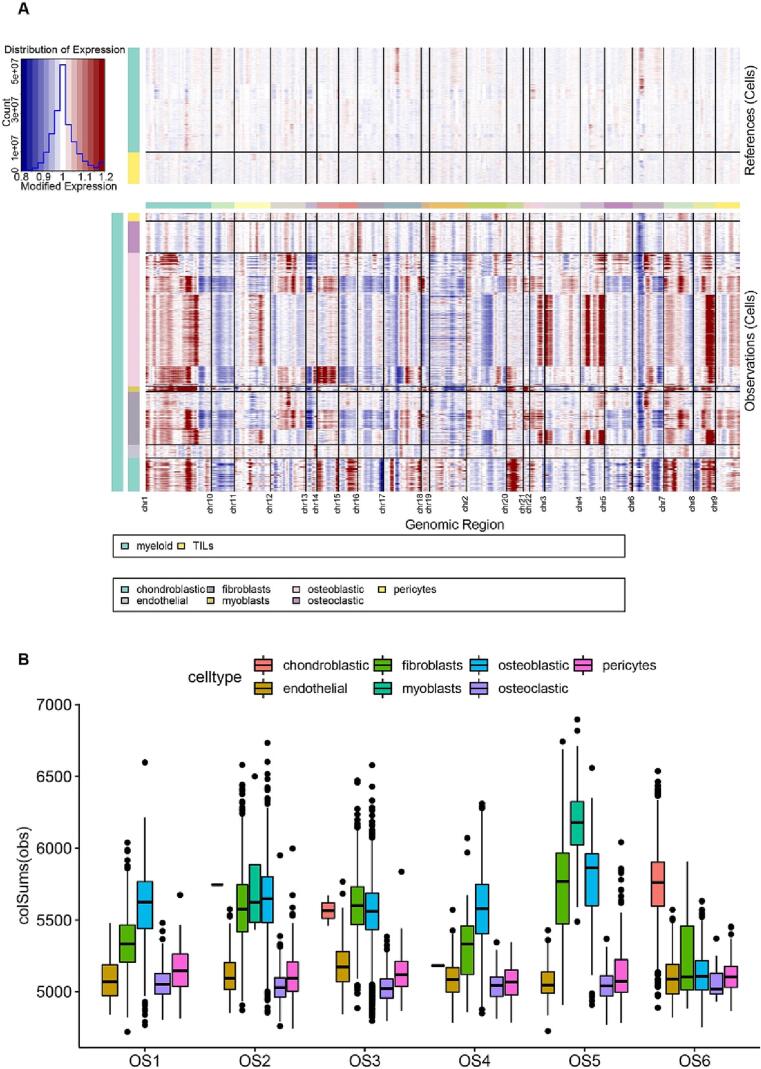
3.2. Copy number variation analysis
Somatic copy-number alterations (SCNA) and structural rearrangements considered to be important features of osteosarcoma.[20] The large-scale chromosomal landscape in OS patients was calculated using reference cells (myeloid cells and TILs). Clustering (Fig. 3A) highlighted three small clusters of non-malignant cells (endothelial cells, pericytes and part of osteoblastic cells), which lack CNVs and highly express markers of specific cell types. The remaining cells formed presumed malignant cells and were associated with CNVs. Our results identified large-scale amplifications and deletions in osteoblastic cells, including the hallmarks of chromosomes 1, 3, 4 and 9 gain (amplification), the chromosomes 2, 6, 10, 15 and 18 regions loss (del) (Fig. 3A), which were consistent with previously reported genomic CNVs[19], [20]. A fraction of fibroblasts also showed similar changes to osteoblastic cells. CNV of Chondroblastic was increased on chr 7, 14, 15, 20 and obviously decreased on chr 10, 6, 3, 4, 5 (Fig. 3A). The results of CNV analysis of different patients showed that osteoblastic, fibroblasts and myoblasts were the three cell types with the highest CNV in five patients, while chondroblastic had the highest CNV in P6 (Fig. 3B).
Fig. 3.
Single-cell copy-number variation in OS. A. The hierarchical heatmap showing large-scale CNVs in OS was calculated using reference cells (myeloid cells and TILs); the red color represents an increased copy number, whereas the blue color represents a decreased copy number. B. CNVs of different cell types in 6 OS patients. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
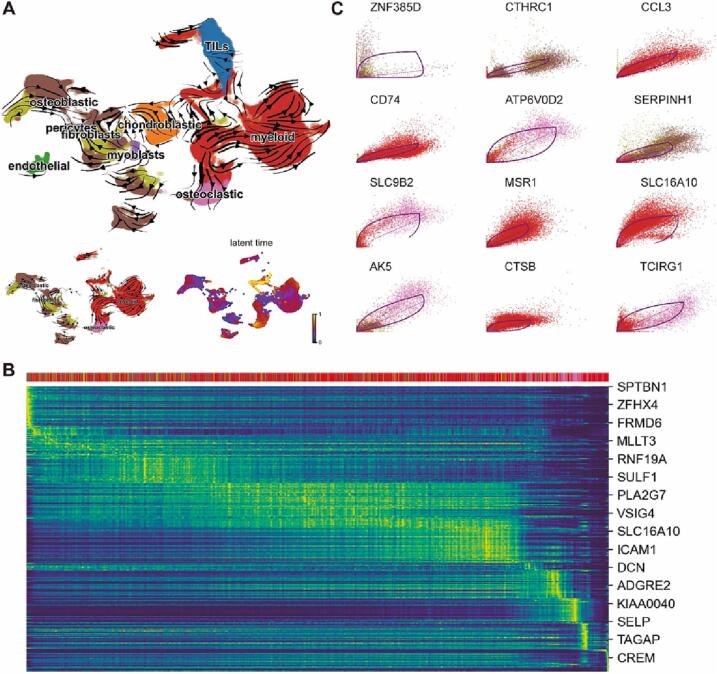
3.3. Trajectory analysis of different cell types in OS
To explore the interrelationship of different cell populations in the OS microenvironment, we analyzed the evolutionary trajectory of different types of cells in OS based on RNA velocity. Osteoclasts are specialized monocyte-macrophage lineage that occupy bone. It has been found that active osteoclasts present at the primary site of OS and plays a vital role in the osteolysis and tumor growth supporting [21]. As expected, our results show that osteoclasts in the OS microenvironment is differentiated from myeloid cells (Fig. 4A), indicating the reliability of this analytical method. RNA velocity gene ranking for different types of cells was listed in supplementary table 2. It‘s worth noting that malignant osteoblastic cells could be trans-differentiated from malignant fibroblast cells (Fig. 4A). Furthermore, the trans-differentiated from fibroblast to osteoblastic cells is not an isolated case, as can be seen from cells of different patient origins (Supplementary Fig. 1).
Fig. 4.
Trajectory analysis of OS single cells. A. RNA Velocity vectors projected onto the UMAP embedding, indicating differentiation directionality. Botom left inset shows four types of cells (osteoblastic, osteoclastic, fibroblasts and myeloid cells). Botom left inset indicates latent time. B. Heatmap showing dynamic RNA Velocity changes in osteoblasts, fibroblasts, myeloid cells and osteoclasts along latent time. C. Identified driver genes with pronounced dynamic behavior. Dashed line indicates steady states. Levels of unspliced mRNA above or below this proportion indicate increasing or decreasing expression of a gene, respectively.
The RNA velocity of the top3 genes in osteoblasts, fibroblasts, myeloid cells and osteoclasts was shown in the phase portraits (Supplementary Fig. 2). Meanwhile, gene expression dynamics resolved along latent time shows a clear cascade of transcription in the top likelihood-ranked genes. Proliferation, differentiation and development related genes, such as SPTBN1, ZFHX4 and FRMD6 (Fig. 4B), were gradually down-regulated along with trajectory differentiation process. Conversely, some well-known immune cells function related factors such as ICAM1, ADGRE2 and TAGAP were upregulated in the process. Furthermore, putative driver genes of trans-differentiation were identified by high likelihoods in a dynamic model [22] (Fig. 4C). Many of the top-ranked genes have been reported to play a crucial role in osteogenic differentiation and development (CTHRC1 and SERPINH1), whereas some of these genes were related to the osteoclast fusion and bone formation (ATP6V0D2 AND TCIRG1).
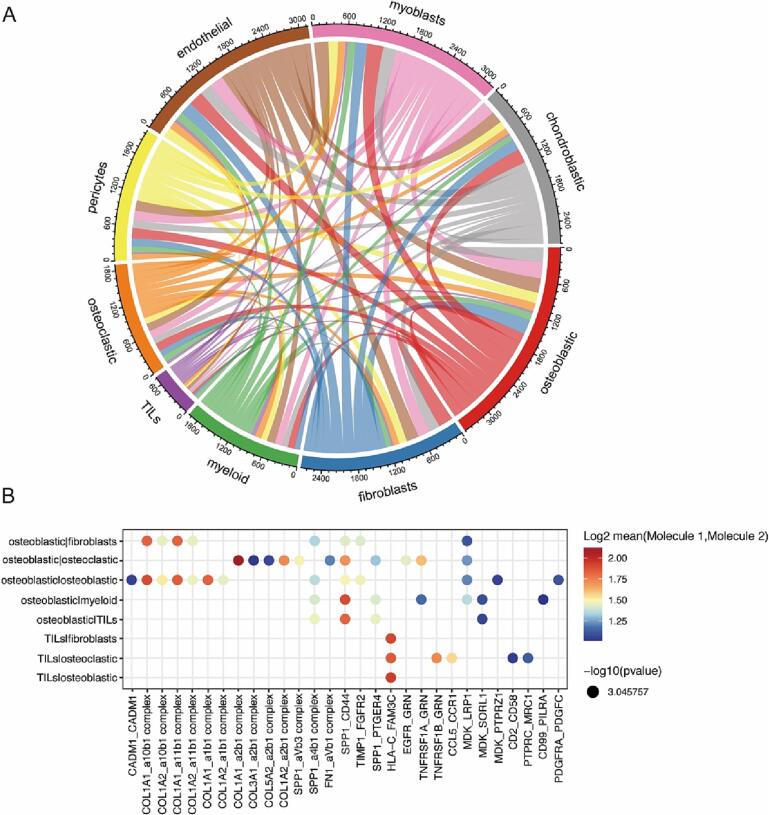
3.4. Ligand/receptor based intercellular communication in OS
In the tumor microenvironment, the communication between different cell types is critical for cancer initiation, progression and therapeutic resistance. To investigate intercellular communications in OS microenvironment, we visualized average expression levels of most abundant ligands and their cognate receptors for each cell type’s ligand-receptor (L-R) pair. A network of potential cell–cell interactions was constructed showing extensive communications between different types of cells. Global overview of the interactomes revealed that osteoblastic cell was the cell type that express the most receptors and ligands, indicating communicate most with other cells (Fig. 5A).
Fig. 5.
Intercellular communication analysis of with osteosarcoma microenvironment. A. Analysis of receptor and ligand interactions between different cell types. Edge thickness is proportional to edge weights. Edge color labels the source cell type. B. Dot plots showing intercellular ligand and receptor pairs in osteoblastic and other cells. The spectrum of color indicates the P value.
We identified multiple ligand-receptor pairs of collagen and integrins in communication involving osteoblastic cell. Osteoblastic cell interacted with osteoclastic via COL1A1-Intergrin α2β1, COL1A2-Intergrin α2β1, interacts with fibroblast via COL1A1-Intergrin α10β1, COL1A1-Intergrin α11β1, which may be associated with osteoblastic cell adhesion, migration, and invasion (Fig. 5B). Also, there is intercellular communication between osteoblastic cells and osteoblastic cells via COL1A1- Intergrin α1β1, COL1A2- Intergrin α11β1 and MDK-PTPRZ1 (Fig. 5B). In addition, we found that some ligand-receptor pairs were present in the communication of osteoblasts and a variety of other cells, such as MDK-LRP1, MDK-SORL1, SPP1 CD44 and SPP1-PTGER4 (Fig. 5B). Furthermore, we analyzed the relationship between ligand-receptors co-expression in OS and patients prognosis. OS patients were divided into four groups by gene expression: ligandhigh receptorhigh, ligandhigh receptorlow, ligandlow receptorhigh and ligandlow receptorlow. Interestingly, 15 ligand-receptors pairs co-expression were found to be associated with patient prognosis (p less than 0.05). Mostly, ligandhigh receptor high patients had the longest survival time and ligandlow receptorlow patients had a poorer prognosis (Fig. 6 and supplementary table 3), such as EGFR-GRN and MDK-SORL1 (Fig. 6 D and F). These results suggest that communication between osteoblastic cells and other cells is a key factor contributing to OS tumor progression and affecting patient prognosis.
Fig. 6.
Prognostic relevance of osteoblastic cell-associated Ligand/receptor pairs expression to OS patients. A. Prognostic analysis of ligand and receptor pairs co-expression in osteosarcoma. P values in bold indicate significant, less than0.05. B-G. Survival curves of ligand and receptor pairs in osteosarcoma.
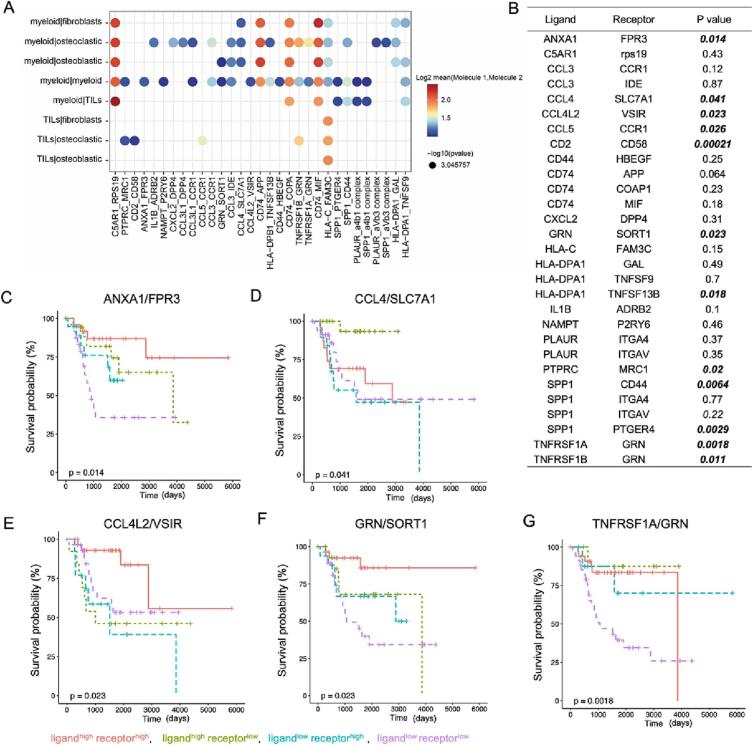
We further analyzed myeloid cells related intercellular communication, as they are the highest proportion of cells in the OS microenvironment. Myeloid cells were found to interact with a variety of other cell types, particularly osteoblasts, in addition to communicating closely with the myeloid cells themselves (Fig. 7A). Among the ligands and receptor pairs with significant results in the analysis, about half were chemokines (CCL3, CCL4, CCL4L2) and MHC related molecules (CD74, HLA-C, HLA-DPA1) and their partners (Fig. 7A), which matched the main immunological functions of myeloid cells. Interactions analysis revealed that communication among C5AR1-RPS19, CD74-MIF/APP/COPA and HLA-C-FAM3C was extensive (Fig. 7A), in agreement with previous findings [23], [24]. These interactions may be associated with the formation of a suppressive immune microenvironment. In addition, we observed a number of ligand-receptor pairs were overlap with osteoclast-associated intercellular communication, including SPP1-CD44, SPP1 ITGA4, SPP1-ITGAV, SPP1-PTGER4, TNFRSF1A-GRN, and TNFRSF1B-GRN. Survival analysis showed that 13 pairs of myeloid cell-associated ligands and receptor pairs were related with prognosis of OS patients (Fig. 7B-G). CCL4-SLC7A1 was found to mediate communication between myeloid and osteoblasts, osteoclasts and fibroblasts. Patients with high expression of CCL4 and low level of SLC7A1 had the best prognosis (Fig. 7D). Potential intercommunications between GRN and SORT1 were found in myeloid and myeloid cells, myeloid and osteoblastic cells. OS patients with high expression of both GRN and SORT1 had better prognosis compared with other groups (Fig. 7F). These results suggest that myeloid cells have multiple important roles in maintaining the complex regulatory network of the tumor microenvironment.
Fig. 7.
Cellular communication of myeloid cells in the OS microenvironment. A. Dot plots showing intercellular ligand and receptor pairs in myeloid cells and other cell types. The spectrum of color indicates the P value. B. Prognostic analysis of ligand and receptor pairs co-expression in osteosarcoma. P values in bold indicate significant. C-G. Survival curves of indicated ligand and receptor pairs co-expression in osteosarcoma.
Furthermore, we found that CD2-CD58, PTPRC-MCR1, and CCL5-CCR1 mediated the interaction between TILs and osteoclasts, and correlated significantly with patient prognosis, with patients with low expression of both receptors and ligands having the worst prognosis (Supplementary Fig. 3). These molecules have been reported to be associated with tumor immune escape and the formation of a suppressive tumor microenvironment [25], [26], [27].
3.5. Differences in intercellular communication in the tumor microenvironment of OS patients receiving chemotherapy or not
It is now generally accepted that chemotherapy and immunotherapy cause a remodeling of the tumor immune microenvironment. To explore whether intercellular communication in the tumor microenvironment of chemotherapy received OS patients differs from treatment naive patients, we further compared and analyzed data of 7 primary OS patients in GSE152048. Most of the above results in this study are in agreement with those obtained by Yan Zhou and his colleagues [19]. Their data showed that the highest percentage in tumor of these patient was also osteoblastic and myeloid cells. Pearson’s correlation between fibroblast and proliferated osteoblastic cells was 0.94, and correlation between fibroblast and osteoblastic cells is 0.85 based on the average gene-expression profiles of the top 3000 highly variable genes among the clusters. These reflect the reliability of the methodology and data of this study.
Interestingly, we found intercellular communication differed in tumors of treatment naïve patients and chemotherapy received patients. We obtained 30 L-R pairs of myeloid communication in treatment naive patients while only 18 in the patients receiving chemotherapy (Fig. 8A and Supplementary Fig. 4). Eight L-R pairs with particularly significant P values were overlapped, including C5AR1-RPS19, CD74-APP, CD74-COPA, CD74-MIF, HLA-C-FAM3C. These L/R pairs were involved in the communication between cells of myeloid origin and almost all other cells Supplementary Fig. 4). In addition, the number of statistically significant L-R pairs associated with osteoblastic cells doubled in patients receiving chemotherapy (Fig. 8B and Supplementary Fig. 5), with a large increase in collagen-integrin families. These data suggest that chemotherapy may cause a decrease in immune cell communication and an increase in cellular communication of malignant osteoblastic cells in OS microenvironment, providing new support for chemotherapy-induced remodeling of the tumor microenvironment.
Fig. 8.
Effect of chemotherapy on intercellular communication in the OS. A. Cellular communication L-R pairs of myeloid cells. B. Cellular communication L-R pairs of osteoblastic cells. Blue represents L-R pairs of OS patients without chemotherapy, red represents patients who received chemotherapy, and gray in the middle indicates the overlap of the two groups. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Numerous studies have shown that intercellular communications and signaling transduction between tumor cells and surrounding cells play an important role in tumor fate determination[24], [28], [29]. In the OS microenvironment, osteoclasts have been shown to promote OS cell proliferation and are associated with OS-induced osteolysis[30]; endothelial has been found to be closely associated with tumor angiogenesis and metastasis [31]; myeloid-derived M2-type macrophages usually directly inhibit the anti-tumor immune function of t cells [32]; and fibroblasts are capable of secreting collagen to mediate OS progression[33]. However, given the genomic complexity and microenvironmental heterogeneity, data on immunotherapy or targeting treatment have not been encouraging[34]. Recent advances in single-cell sequencing technologies have allowed us to further explore intercellular communication relationships in the OS microenvironment. Through integrative analysis of scRNA-seq data from treatment naive OS samples, we identified fibroblasts as a possible source of malignant osteoblasts and provided. Our data also provide important ligand/receptor pairs involved in intercellular communication in the OS microenvironment, some of which are highly correlated with receptor expression levels and prognosis of OS patients. These key molecules for intercellular communication help us to better understand the mechanisms of OS progression and may become potential targets for new OS therapeutic strategies.
It's well known that multiple important cell types in mammalian bone tissue have a mesenchymal origin, including fibroblasts, osteoblasts, chondroblasts. These cells can undergo transdifferentiation under specific conditions or environments, such as transdifferentiation of chondrocytes into osteoblasts [19], [35], transdifferentiation of fibroblasts into osteoblasts[36], [37], [38]. I-Ning E Wang et al.[39] found that the interaction of osteoblasts and fibroblasts lead to cell trans-differentiation and fibrocartilage formation in a vitro co-culture system. In this study, we found that a fraction of malignant osteoblasts in the OS tumor microenvironment could be transformed from fibroblasts by CNV and trajectory analysis of single cell sequencing data. It's worth noting that our data showed there was a strong correlation between osteoblast and fibroblast in OS. Similar results have been reported by Yan Zhou et al [19]. In this study, osteoblastic cells were characterized with high COL1A1, CDH11 and RUNX2 expression, the fibroblasts were defined by COL1A1, LUM and DCN. These genes are not absolutely negative or positive expression, e.g. COL1A1, CDH11 and RUNX2 are expressed on both osteoblasts and fibroblasts, while LUM and DCN are expressed on osteoblasts, only with differences in expression intensity. It is possible that some marker gene moderately expressed cells, or cells of intermediate status are inaccurately categorized. Therefore, these results need to be further validated by more accurate cell lineage tracing experiments, as the data analysis in this study cannot fully exclude the effect of a few cell types that failed to cluster accurately, such as MSC, fibroblast-like cells. There may be a neoplastic cell population with varying phenotype in OS.
Integrins, ubiquitous expressed heterodimeric transmembrane glycoprotein receptors, are a class of cell adhesion and signaling proteins involved in a variety of physiological and pathological processes including embryonic development[40], tumor cell growth and metastasis[41], bone resorption[42], and cellular responses to mechanical stress[43]. Currently, drugs targeting integrins have been extensively developed in areas including cardiovascular diseases, dry eye diseases, autoimmune diseases and inflammatory diseases [44], [45], [46], [47]. Abnormal expression of collagen and integrins and the interaction between the two have been shown to be involved in the development, cell adhesion, migration, and invasion of osteosarcoma [48], [49], [50]. In the treatment naïve OS microenvironment, we identified 10 ligand-receptor pairs of collagen and integrins involved in communication of osteoblasts, which increased to 36 in post-chemotherapy patients, reflecting the responsiveness of osteoblasts-associated cellular communication to chemotherapy.
Myeloid cells are important components of the OS microenvironment, and our data show that myeloid cells account for approximately half of the total cell population. We found that the intercellular communication of myeloid cells in treatment naïve patients was mainly dependent on proinflammatory chemokines and MHC-related molecules, suggesting that the main role of myeloid cells in the OS immune microenvironment is the recruitment of immune cells and antigen presentation. However, in the microenvironment of OS patients after chemotherapy, the number of L-R pairs decreased substantially and no chemokines related L-R pairs were found for myeloid cells communication. The main function of CCL3 and CCL4 is to mediate the recruitment of macrophages in the tumor microenvironment, and in this study, CCL3-CCR1, CCL3-IDE, CCL4-SLC7A1 interaction occurred between myeloid and myeloid cells, myeloid and osteoblastic cells. In addition, there are some receptor-ligands in our results that have not been reported in osteosarcoma, such as GRN-SORT1, TNFRSF1A-GRN, and TNFRSF1B-GRN. GRN regulates lysosomal biogenesis, neurons survival and axonal outgrowth[51], [52], and its abnormal function has been reported in several neurological diseases such as frontotemporal dementia [53]and neuronal ceroid lipofuscinosis[52]. GRN/TNFRSF1A has been reported to be associated with macrophage infiltration in gastric cancer[54], while its role in osteosarcoma needs further experimental elucidation.
In summary, we have analyzed the heterogeneity of the tumor microenvironment in OS, and identified the trans-differentiation of malignant osteoblastic cells from fibroblast cells. There are differences in intercellular communication in the tumor microenvironment of OS patients who received or did not receive chemotherapy. The study has also provided intercellular communicating ligand-receptor pairs that associated with OS survival, which will lay the foundation for future research studies on therapy of OS.
Funding
This study was supported by Suzhou Medical Innovation Applied Research Project (SKYD2022090) and the Youth Project of Shanghai Municipal Health and Family Planning Commission (20184Y0141).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbo.2023.100493.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Longo D.L., Meltzer P.S., Helman L.J. New horizons in the treatment of osteosarcoma. N. Engl. J. Med. 2021;385(22):2066–2076. doi: 10.1056/NEJMra2103423. [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L., Troisi R.J., Savage S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao X., Wu Q., Gong X., Liu J., Ma Y. Osteosarcoma: a review of current and future therapeutic approaches. Biomed. Eng. Online. 2021;20(1):24. doi: 10.1186/s12938-021-00860-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14(11):722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 5.Finck A.V., Blanchard T., Roselle C.P., Golinelli G., June C.H. Engineered cellular immunotherapies in cancer and beyond. Nat. Med. 2022;28(4):678–689. doi: 10.1038/s41591-022-01765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski T.J., Biederstadt A., Rezvani K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat. Rev. Cancer. 2022;22(10):557–575. doi: 10.1038/s41568-022-00491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morad G., Helmink B.A., Sharma P., Wargo J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell. 2021;184(21):5309–5337. doi: 10.1016/j.cell.2021.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes D.J., Dutton P., Bruland Ø., Gelderblom H., Faleti A., Bühnemann C., van Maldegem A., Johnson H., Poulton L., Love S., Tiemeier G., van Beelen E., Herbschleb K., Haddon C., Billingham L., Bradley K., Ferrari S., Palmerini E., Picci P., Dirksen U., Strauss S.J., Hogendoorn P.C.W., Buddingh E., Blay J.-Y., Cleton-Jansen A.M., Hassan A.B. Outcomes from a mechanistic biomarker multi-arm and randomised study of liposomal MTP-PE (Mifamurtide) in metastatic and/or recurrent osteosarcoma (EuroSarc-Memos trial) BMC Cancer. 2022;22(1) doi: 10.1186/s12885-022-09697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nørregaard K.S., Jürgensen H.J., Gårdsvoll H., Engelholm L.H., Behrendt N., Søe K. Osteosarcoma and metastasis associated bone degradation-A tale of osteoclast and malignant cell cooperativity. Int. J. Mol. Sci. 2021;22(13):6865. doi: 10.3390/ijms22136865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C., Tian Y.e., Zhao F., Chen Z., Su P., Li Y.u., Qian A. Bone microenvironment and osteosarcoma metastasis. Int. J. Mol. Sci. 2020;21(19):6985. doi: 10.3390/ijms21196985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanotti S., Canalis E. Notch signaling and the skeleton. Endocr. Rev. 2016;37(3):223–253. doi: 10.1210/er.2016-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura S. Single-cell genomics to understand disease pathogenesis. J. Hum. Genet. 2021;66(1):75–84. doi: 10.1038/s10038-020-00844-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gohil S.H., Iorgulescu J.B., Braun D.A., Keskin D.B., Livak K.J. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021;18(4):244–256. doi: 10.1038/s41571-020-00449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L., Qin S., Si W., Wang A., Xing B., Gao R., Ren X., Wang L.i., Wu X., Zhang J.i., Wu N., Zhang N., Zheng H., Ouyang H., Chen K., Bu Z., Hu X., Ji J., Zhang Z. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374(6574) doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Chen H., Mo H., Hu X., Gao R., Zhao Y., Liu B., Niu L., Sun X., Yu X., Wang Y., Chang Q., Gong T., Guan X., Hu T., Qian T., Xu B., Ma F., Zhang Z., Liu Z. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell. 2021;39(12):1578–1593.e8. doi: 10.1016/j.ccell.2021.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M, et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 12(3): 670-691. [DOI] [PMC free article] [PubMed]
- 17.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17(2):159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 18.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A., Fan J., Borm L.E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P.V. RNA velocity of single cells. Nature. 2018;560(7719):494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Yang D., Yang Q., Lv X., Huang W., Zhou Z., Wang Y., Zhang Z., Yuan T., Ding X., Tang L., Zhang J., Yin J., Huang Y., Yu W., Wang Y., Zhou C., Su Y., He A., Sun Y., Shen Z., Qian B., Meng W., Fei J., Yao Y., Pan X., Chen P., Hu H. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-20059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayles LC, Breese MR, Koehne AL, Leung SG, Lee AG, Liu HY, et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019, 9(1): 46-63. [DOI] [PMC free article] [PubMed]
- 21.Akiyama T, Dass CR, Choong PF. Novel therapeutic strategy for osteosarcoma targeting osteoclast differentiation, bone-resorbing activity, and apoptosis pathway. Mol Cancer Ther. 2008, 7(11): 3461-3469. [DOI] [PubMed]
- 22.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38(12):1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 23.Ye C., Zhu J., Wang J., Chen D., Meng L., Zhan Y., Yang R., He S., Li Z., Dai S., Li Y.i., Sun S., Shen Z., Huang Y., Dong R., Chen G., Zheng S. Single-cell and spatial transcriptomics reveal the fibrosis-related immune landscape of biliary atresia. Clin. Transl. Med. 2022;12(11):e1070. doi: 10.1002/ctm2.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JJ, Bernard V, Semaan A, Monberg ME, Huang J, Stephens BM, et al. Elucidation of Tumor-Stromal Heterogeneity and the Ligand-Receptor Interactome by Single-Cell Transcriptomics in Real-world Pancreatic Cancer Biopsies. Clin Cancer Res. 2021, 27(21): 5912-5921. [DOI] [PMC free article] [PubMed]
- 25.Zhang Y., Liu Q., Yang S., Liao Q. CD58 immunobiology at a glance. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.705260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Li M., Wei R., Wu J. Identification and functional analysis of EPOR(+) tumor-associated macrophages in human osteosarcoma lung metastasis. J. Immunol. Res. 2020;2020:1–14. doi: 10.1155/2020/9374240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S.-W., Wu H.-H., Liu S.-C., Wang P.-C., Ou W.-C., Chou W.-Y., Shen Y.-S., Tang C.-H., Tsilibary E.C. CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PLoS One. 2012;7(4):e35101. doi: 10.1371/journal.pone.0035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato A., Rahman N.I.A., Shimizu A., Ogita H. Cell-to-cell contact-mediated regulation of tumor behavior in the tumor microenvironment. Cancer Sci. 2021;112(10):4005–4012. doi: 10.1111/cas.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreps L.M., Addison C.L. Targeting intercellular communication in the bone microenvironment to prevent disseminated tumor cell escape from dormancy and bone metastatic tumor growth. Int. J. Mol. Sci. 2021;22(6):2911. doi: 10.3390/ijms22062911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohba T., Cole H.A., Cates J.MM., Slosky D.A., Haro H., Ando T., Schwartz H.S., Schoenecker J.G. Bisphosphonates inhibit osteosarcoma-mediated osteolysis via attenuation of tumor expression of MCP-1 and RANKL. J. Bone Miner. Res. 2014;29(6):1431–1445. doi: 10.1002/jbmr.2182. [DOI] [PubMed] [Google Scholar]
- 31.Yang J., Hu Y., Wang L., Sun X., Yu L., Guo W. Human umbilical vein endothelial cells derived-exosomes promote osteosarcoma cell stemness by activating Notch signaling pathway. Bioengineered. 2021;12(2):11007–11017. doi: 10.1080/21655979.2021.2005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Q., Shi H., Liu F. CD163(+) M2-type tumor-associated macrophage support the suppression of tumor-infiltrating T cells in osteosarcoma. Int. Immunopharmacol. 2016;34:101–106. doi: 10.1016/j.intimp.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Liu Z., Yang X., Lu W., Chen Y., Lin Y., Wang J., Lin S., Yun J.-P. H3K27 acetylation activated-COL6A1 promotes osteosarcoma lung metastasis by repressing STAT1 and activating pulmonary cancer-associated fibroblasts. Theranostics. 2021;11(3):1473–1492. doi: 10.7150/thno.51245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahiro K., Matsumoto Y. Immunotherapy for osteosarcoma. Hum. Vaccin. Immunother. 2021;17(5):1294–1295. doi: 10.1080/21645515.2020.1824499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin X., Jiang Q., Nagano K., Moriishi T., Miyazaki T., Komori H., Ito K., Mark K.V.D., Sakane C., Kaneko H., Komori T., Long F. Runx2 is essential for the transdifferentiation of chondrocytes into osteoblasts. PLoS Genet. 2020;16(11):e1009169. doi: 10.1371/journal.pgen.1009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi A., Nagata M., Gupta A., Matsushita Y., Yamaguchi T., Mizuhashi K., Maki K., Ruellas A.C., Cevidanes L.S., Kronenberg H.M., Ono N., Ono W. Autocrine regulation of mesenchymal progenitor cell fates orchestrates tooth eruption. Proc. Natl. Acad. Sci. U. S. A. 2019;116(2):575–580. doi: 10.1073/pnas.1810200115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu Z.F., Chiu J., Lee L.R., Schindeler A., Jackson M., Ramaswamy Y., Dunstan C.R., Hogg P.J., Zreiqat H. Reprogramming of human fibroblasts into osteoblasts by insulin-like growth factor-binding protein 7. Stem Cells Transl. Med. 2020;9(3):403–415. doi: 10.1002/sctm.19-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cayami F.K., Claeys L., de Ruiter R., Smilde B.J., Wisse L., Bogunovic N., Riesebos E., Eken L., Kooi I., Sistermans E.A., Bravenboer N., Pals G., Faradz S.M.H., Sie D., Eekhoff E.M.W., Micha D. Osteogenic transdifferentiation of primary human fibroblasts to osteoblast-like cells with human platelet lysate. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-18512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang I.-N., Shan J., Choi R., Oh S., Kepler C.K., Chen F.H., Lu H.H. Role of osteoblast-fibroblast interactions in the formation of the ligament-to-bone interface. J. Orthop. Res. 2007;25(12):1609–1620. doi: 10.1002/jor.20475. [DOI] [PubMed] [Google Scholar]
- 40.Darribere T., Skalski M., Cousin H.L., Gaultier A., Montmory C., Alfandari D. Integrins: regulators of embryogenesis. Biol. Cell. 2000;92(1):5–25. doi: 10.1016/s0248-4900(00)88760-2. [DOI] [PubMed] [Google Scholar]
- 41.Hamidi H., Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat. Rev. Cancer. 2018;18(9):533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartman G.D., Duggan M.E. alpha(v)beta(3) Integrin antagonists as inhibitors of bone resorption. Expert Opin. Investig. Drugs. 2000;9(6):1281–1291. doi: 10.1517/13543784.9.6.1281. [DOI] [PubMed] [Google Scholar]
- 43.Winograd-Katz S.E., Fassler R., Geiger B., Legate K.R. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014;15(4):273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 44.Slack R.J., Macdonald S.J.F., Roper J.A., Jenkins R.G., Hatley R.J.D. Emerging therapeutic opportunities for integrin inhibitors. Nat. Rev. Drug Discov. 2022;21(1):60–78. doi: 10.1038/s41573-021-00284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lal H., Guleria R.S., Foster D.M., Lu G., Watson L.E., Sanghi S., et al. Integrins: novel therapeutic targets for cardiovascular diseases. Cardiovasc. Hematol. Agents Med. Chem. 2007;5(2):109–132. doi: 10.2174/187152507780363223. [DOI] [PubMed] [Google Scholar]
- 46.Gubatan J., Keyashian K., Rubin S.J.S., Wang J., Buckman C.A., Sinha S. Anti-integrins for the treatment of inflammatory bowel disease: Current evidence and perspectives. Clin. Exp. Gastroenterol. 2021;14:333–342. doi: 10.2147/CEG.S293272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mrugacz M., Bryl A., Falkowski M., Zorena K. Integrins: An important link between angiogenesis, inflammation and eye diseases. Cells. 2021;10(7):1703. doi: 10.3390/cells10071703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei D., Li C., Ye J., Xiang F., Liu J. Extracellular collagen mediates osteosarcoma progression through an integrin alpha2beta1/JAK/STAT3 signaling pathway. Cancer Manag. Res. 2020;12:12067–12075. doi: 10.2147/CMAR.S273466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vihinen P., Riikonen T., Laine A., Heino J. Integrin alpha 2 beta 1 in tumorigenic human osteosarcoma cell lines regulates cell adhesion, migration, and invasion by interaction with type I collagen. Cell Growth Differ. 1996;7(4):439–447. [PubMed] [Google Scholar]
- 50.Gvozdenovic A., Boro A., Meier D., Bode-Lesniewska B., Born W., Muff R., et al. Targeting alphavbeta3 and alphavbeta5 integrins inhibits pulmonary metastasis in an intratibial xenograft osteosarcoma mouse model. Oncotarget. 2016;7(34):55141–55154. doi: 10.18632/oncotarget.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beel S., Moisse M., Damme M., De Muynck L., Robberecht W., Van Den Bosch L., Saftig P., Van Damme P. Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum. Mol. Genet. 2017;26(15):2850–2863. doi: 10.1093/hmg/ddx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka Y., Suzuki G., Matsuwaki T., Hosokawa M., Serrano G., Beach T.G., et al. Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum. Mol. Genet. 2017;26(5):969–988. doi: 10.1093/hmg/ddx011. [DOI] [PubMed] [Google Scholar]
- 53.Greaves C.V., Rohrer J.D. An update on genetic frontotemporal dementia. J. Neurol. 2019;266(8):2075–2086. doi: 10.1007/s00415-019-09363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao L., Wang W., Niu P., Luan X., Zhao D., Chen Y. The molecular mechanisms of CTHRC1 in gastric cancer by integrating TCGA, GEO and GSA datasets. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.