Figure 1.
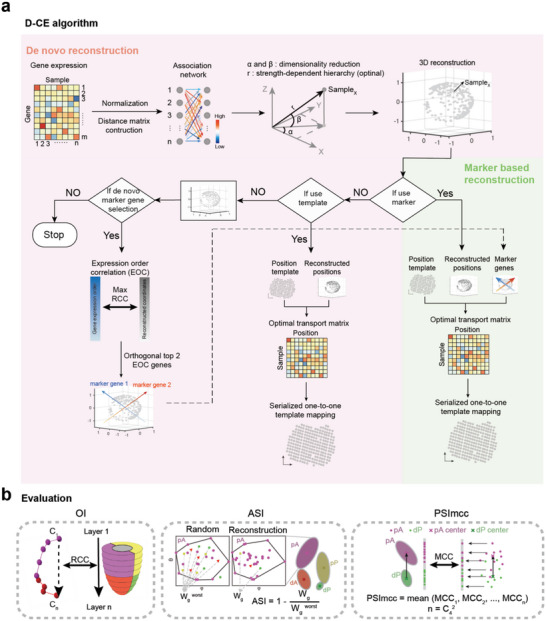
D‐CE method of spatial reconstruction of 3D localizations. a) Overview of spatial structure reconstruction by D‐CE, which consists of angular and radial reconstruction. Angular reconstruction is based on dimensionality reduction by singular value decomposition (SVD), radial reconstruction is based on the strength‐dependent hierarchy. Pearson distance and SQRT (square root of each element of the matrix) were applied to all expressed genes to construct an association network for D‐CE. Maximum EOC (expression order correlation) were calculated for all expressed genes in all directions by rotating the reconstructed structure at 12° per step in two orthogonal directions, respectively. If a position template is available with or without using marker genes, the D‐CE reconstructed coordinates were serially one‐to‐one mapped to the template after optimal transport to D‐CE‐t reconstructions. The top EOC genes on the 2 and 3 orthogonal directions are use as marker gene 1, 2, and 3 for 2D and 3D template fitting, respectively. b) Then ordering index (OI), angular separation index (ASI), and projection separability index of Matthews correlation coefficient (PSImcc) to the original sample layer order were used to assess the accuracy of spatial reconstruction of samples’ positions.
