Figure 2.
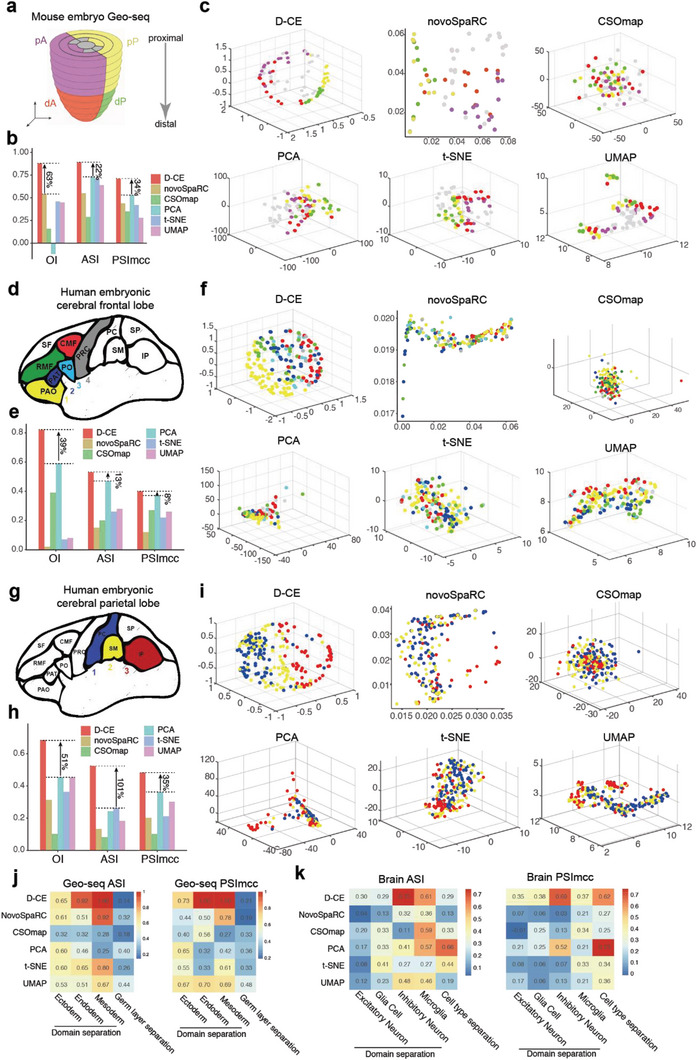
Reconstruction of spatial domain labels from oligo or single cell RNA‐seq data with by D‐CE, novoSpaRC, CSOmap, PCA, t‐SNE, and UMAP. a) Illustration of 4 spatial domains pA, dA, pP, and dP in mouse embryo E7.5. b) Barplot of OI, ASI, and PSImcc for D‐CE, novoSpaRC, CSOmap, PCA, t‐SNE, and UMAP reconstructions. c) Reconstructed structure of D‐CE, novoSpaRC, CSOmap, PCA, t‐SNE, and UMAP. d–f) The same layout as panel (a) to (c) for human embryonic frontal lobe. Six regions from frontal lobe, CMF (caudal‐middle‐frontal), PRC (precentral), PAO (pars orbitalis), PO (pars opercularis), RMF (rostral‐middle‐frontal), and PAT (pars triangulars), are used as domain labels of the scRNA‐seq data. As only microglia cells contain >3 cells per region, they are the only cell type used for reconstruction. OI is calculated between the known domain order and the reconstructed coordinates of PAO, PAT, PO, and PRC (ordered from 1–4). g–i) The same layout as panel (a–c) for human embryonic parietal lobe. 3 regions from parietal lobe (PC (postcentral), SM (supra‐maginal), and IP (inferior parietal)) as domain labels. OI is calculated between the known order and the reconstructed coordinates of PC, SM, and IP (ordered from 1 to 3). For panels (b, e, and h) the percentage of improvement by D‐CE over the second‐best method is labeled for each index. j) ASI (left panel) and PSImcc (right panel) for spatial domain separation and reconstruction of Geo‐seq samples from the same germ layer, as indicated (≈1–3 columns), or for the separation of different germ layers (the last column) by each reconstruction method. k) ASI (left panel) and PSImcc (right panel) for spatial domain separation and reconstruction of single cells in the brain parietal region scRNA‐seq data in panel (g), within each cell type that contains >3 cells per region, as indicated (≈1–4 columns), or the separation of different cell types (the last column).
