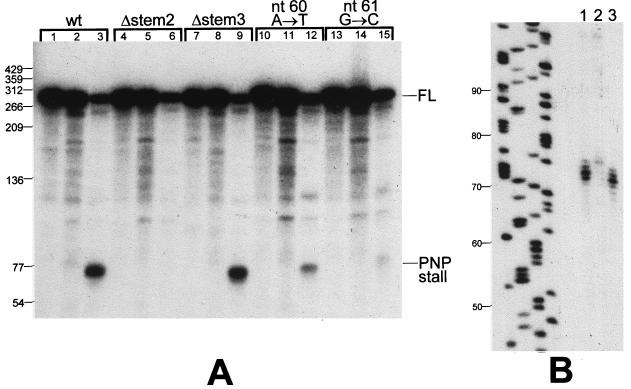
FIG. 5.
In vitro analysis of PNPase processing. (A) Low-resolution denaturing gel showing processing of labeled SP82 RNA incubated in the presence of a BG218 protein extract. The RNA substrate is indicated at the top of each set (Δ, deletion mutant). The three lanes in each set contained a control in which RNA processing was inactivated by the immediate addition of phenol-chloroform (lanes 1, 4, 7, 10, and 13), the mixture without Pi (lanes 2, 5, 8, 11, and 14), and the mixture with Pi (lanes 3, 6, 9, 12, and 15). (B) High-resolution denaturing gel showing the migration of the PNPase processing product from the wild-type (lane 1) and stem 3 deletion mutant (lane 3) substrates. The leftmost lanes show a sequencing reaction used as a molecular size marker (sizes in nucleotides are indicated at the left). Lane 2 contains RNA that was incubated with the extract in the presence of Pi and 3 mM ATP. The products of this incubation migrated near the top of the gel (not shown), suggesting the presence of polyadenylating activity in the extract. This result is not relevant to the subject of this paper.