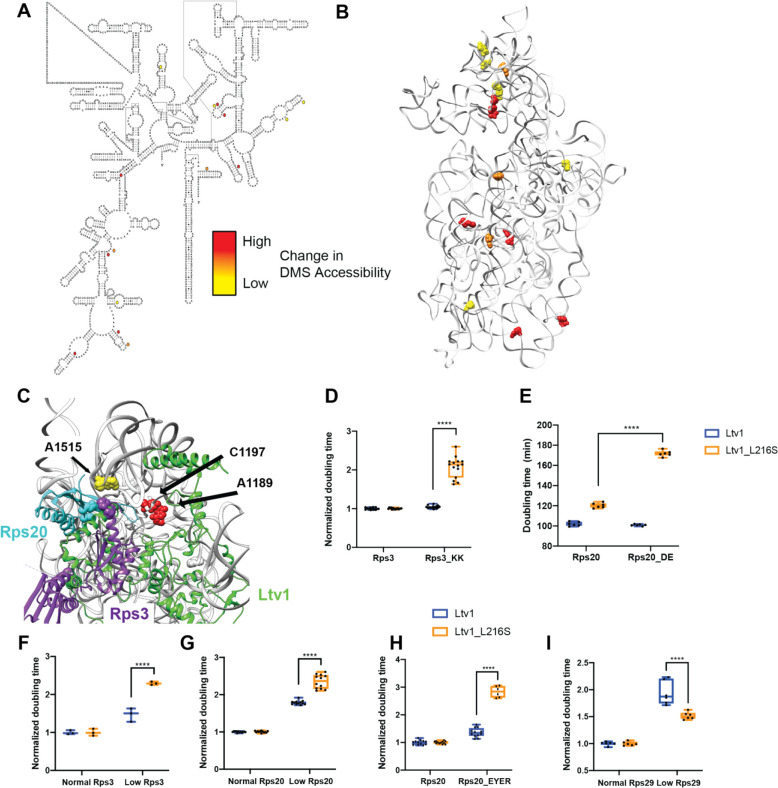
Figure 6: DMS MaPSeq demonstrates perturbations in rRNA folding arising from the Ltv1_L216S mutation.
(A) Secondary structure of 18S rRNA demonstrating residues significantly altered in ribosomes from Ltv1_L216S cells. All residues (except C191) are more accessible in ribosomes from Ltv1_L216S. The extent of the alteration is indicated by yellow, orange or red circles. (B) Residues significantly altered in ribosomes from Ltv1_L216S cells mapped onto the 3D structure of 18S rRNA from pre-40S ribosomes (PDB ID 6FAI). Color scheme as in panel A. (C) Highlight of the composite structure from Figure 3, demonstrating that A1515, A1189 and C1197 are adjacent to residues in Ltv1. A1196 is not resolved in the structure and therefore not shown. Rps20_DE and Rps3_KK are shown in cyan and purple spheres, respectively. The Ltv1L216S mutation demonstrates significant synthetic sick or lethal interactions with Rps3_KK (D) or Rps20_DE (E), respectively. (F) Doubling times (normalized to normal Rps3) for yeast cells expressing either normal or low levels of Rps3 and wt Ltv1 or Ltv1_L216S. Normal levels of Rps3 and Rps20 were obtained by using the TEF2 promoter-driven plasmids. Low levels of Rps3 and Rps20 were obtained by using Tet-promoter-driven plasmids and addition of 0 or 20 ng/ml of doxycycline (dox), for Rps3 and Rps20, respectively. Significance was tested using an unpaired t-test. ****, P<0.0001. (G) Doubling times (normalized to normal Rps20) for yeast cells expressing either normal or low levels of Rps20 and wt Ltv1 or Ltv1_L216S. Significance was tested using an unpaired t-test. ****, P<0.0001. (H) Doubling times (normalized to wt Rps20) for yeast cells expressing either wt Rps20 or Rps20_EYER and wt Ltv1 or Ltv1_L216S. Significance was tested using an unpaired t-test. ****, P<0.0001. (I) Doubling times (normalized to normal Rps29) for yeast cells expressing either normal or low levels of Rps29 and wt Ltv1 or Ltv1_L216S. Low levels of Rps29 were obtained by using a Tet-promoter-driven plasmid, which even without dox addition produces reduced levels of Rps29. Normal levels of Rps29 were produced with TEF2 promoter-driven plasmids. Significance was tested using an unpaired t-test. ****, P<0.0001.