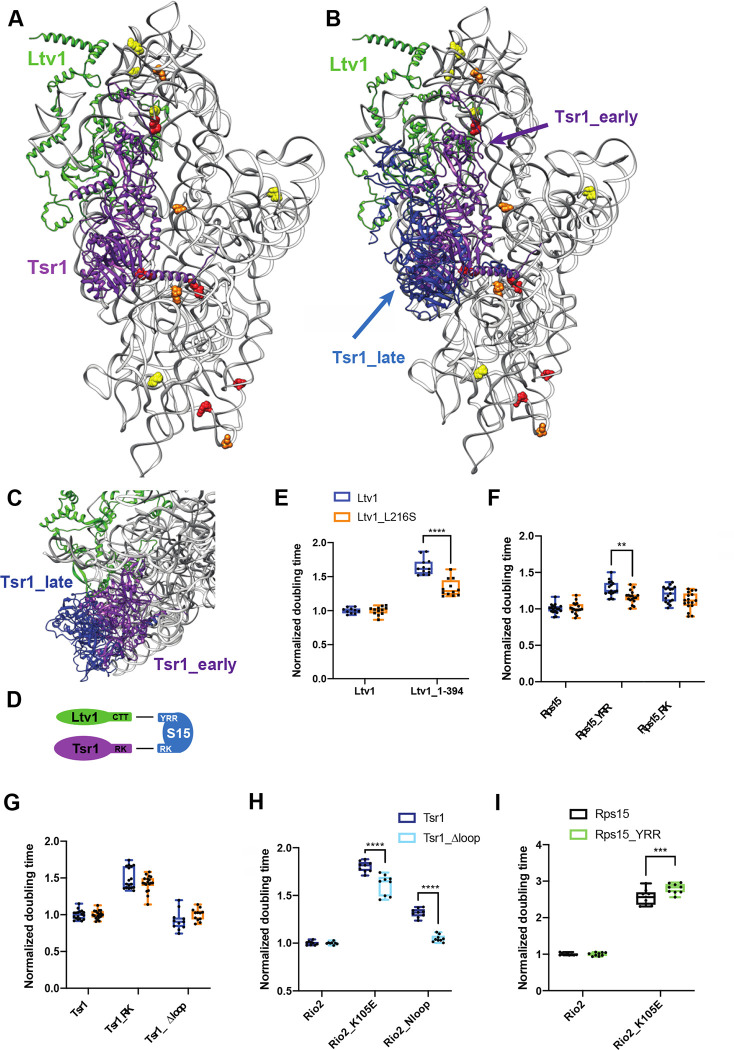
Figure 7: Ltv1 dissociation requires Tsr1 movement to the beak.
(A) Structure of the pre-40S ribosome highlighting residues altered in ribosomes from Ltv1_L216S yeast as determined by DMS-MaPseq (Figure 6A&B) showing Tsr1 and Ltv1 (from PDB ID 6FAI). (B) Structure of the pre-40S ribosome with Ltv1 and Tsr1 in the position from earlier pre-40S (6FAI, purple) and from 80S-like ribosomes (in blue), near the beak (6WDR). 6FAI and 6WDR were overlaid by matching the 18S rRNA. (C) Structural detail of the complex in B, viewed from the top of the subunit, demonstrating that Tsr1_early, but not Tsr1_late blocks binding and dissociation of Ltv1. (D) Summary of the genetic interaction network shown by the data in panel E and F and in Figure S5. (E) Doubling times (normalized to wt Ltv1) for yeast cells expressing either wt Ltv1 or Ltv1_L216S in full length or truncated (at amino acid 394) form. Significance was tested using an unpaired t-test. ****, P<0.0001. (F) Doubling times (normalized to wt Rps15) for yeast cells expressing either wt Rps15, Rps15_YRR or Rps15_RK and wt Ltv1 or Ltv1_L216S. Significance was tested using an unpaired t-test. **, P<0.01. (G) Doubling times (normalized to wt Tsr1) for yeast cells expressing either wt Tsr1 or Tsr1_RK or Tsr1Δloop and wt Ltv1 or Ltv1_L216S. Significance was tested using an unpaired t-test. (H) Doubling times (normalized to wt Rio2) for yeast cells expressing either wt Rio2, Rio2_K105E or Rio2_loop. Significance was tested using an unpaired t-test. ****, P<0.0001. Data for Tsr1_RK and Tsr1_Δhinge are in Figure S5e of [16] (I) Doubling times (normalized to wt Rio2) for yeast cells expressing either wt Rio2, or Rio2_K105E. Significance was tested using an unpaired t-test. ***, P<0.001. Data for Tsr1_RK and Tsr1_Δhinge are in Figure S5g of [16].