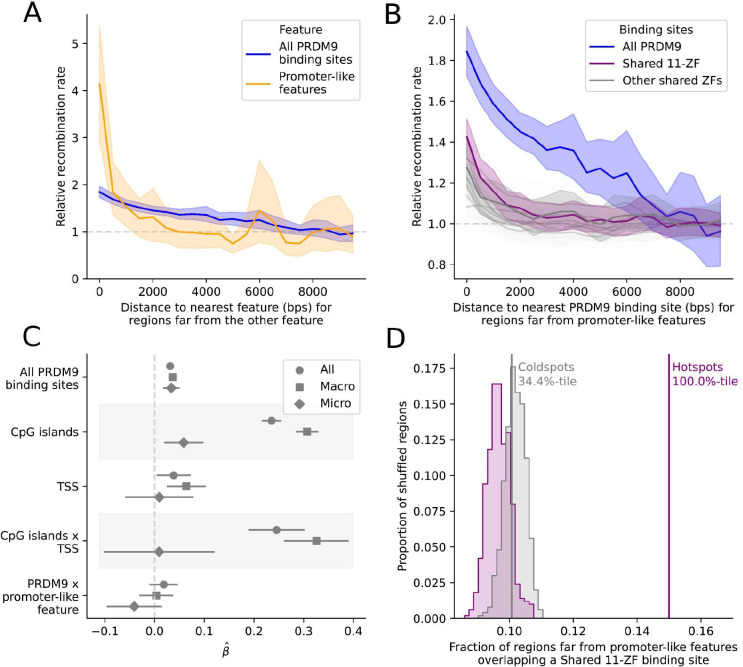
Figure 2: An increase in the population recombination rate is seen around both PRDM9 binding sites and promoter-like features.
A) Mean population recombination rate in 100 bp windows as a function of distance to the nearest predicted PRDM9 binding site (blue) or promoter-like feature (orange). When considering one feature, we condition on windows >10 kb from the other feature; thus, when focusing on predicted PRDM9 binding sites, we only consider windows that are >10 kb from promoter-like features (i.e., TSS or CpG island). The recombination rate is relative to the mean rate 8–10 kb away. Shaded regions represent the central 95% confidence interval obtained by bootstrapping (see Methods). B) Mean population recombination in 100 bp windows as a function of distance from predicted binding sites for sets of zinc fingers shared among PRDM9 alleles. This plot is conditional on the windows being far from promoter-like features. The “Shared 11-ZF” allele shared among five PRDM9 alleles is shown in purple and the set of all PRDM9 alleles, equivalent to the curve in (A), is shown in blue. C) Point estimate and 95% CI for the coefficients of a linear model, in which the response variable is the (log) recombination rate in 1 kb windows (thinned to be 10 kb apart) and the predictors are the (binary) presence or absence of one or more predicted PRDM9 binding sites, TSS, or CpG islands. Covariates include the background recombination rate (1 Mb scale) and GC content (see Methods). Results are reported for data from the autosomes (circles), only scaffolds assigned to macrochromosomes (squares), and only microchromosomes (diamonds). D) Overlap of hotspots (purple) and matched coldspots (gray) far from a promoter-like feature with the predicted binding sites for the Shared 11-ZF allele. The observed values are shown with solid lines. The overlap expected by chance is shown with the shaded distribution and is based on 500 replicates, in which each hotspot was placed at random within 5 Mb of the original location conditional on there not being a gap in the genome sequence (see Methods). We note that while hotspots and coldspots are matched for base composition (see Methods), that need no longer be the case once we condition on them laying far from a promoter-like feature, driving the slight difference between the null distributions.