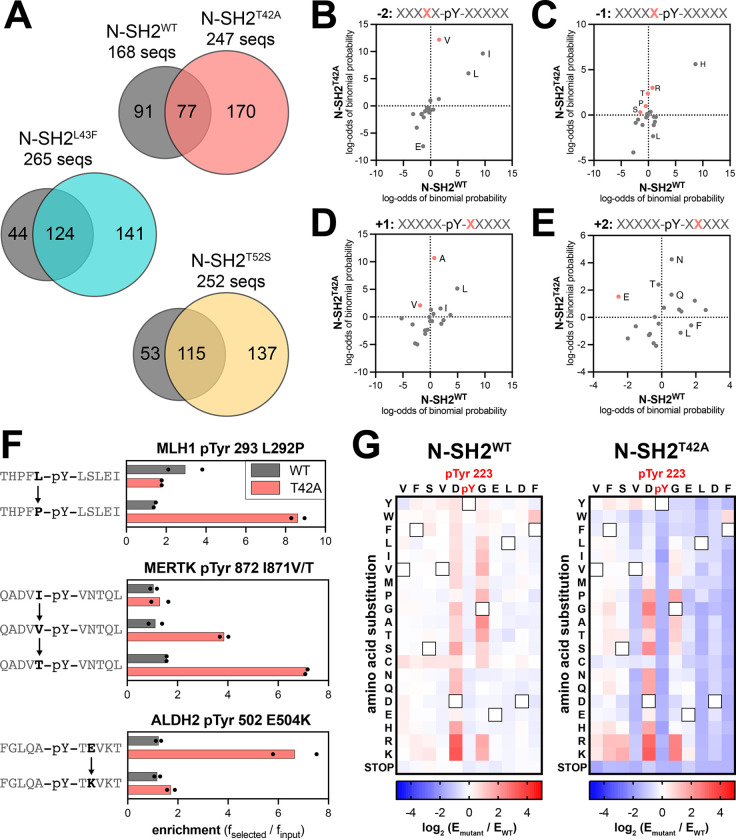
Figure 4. Analysis of enriched sequence features reveals specificity differences at several positions.
(A) Overlap in sequences enriched by each N-SH2 domain above an enrichment score cutoff of 3.2. (B) Log-transformed probabilities of amino acid enrichment at the −2 position relative to the pTyr residue, derived from peptide display screens with N-SH2WT and N-SH2T42A. (C) Same as panel (B), except at the −1 position. (D) Same as panel (B), except at the +1 position. (E) Same as panel (B), except at the +2 position. (F) Enrichment scores for representative sets of peptides from the pTyr-Var Library screens that highlight different sequence preferences for N-SH2WT and N-SH2T42A. In each sub-panel, a pair or trio of peptides are shown that differ by one amino acid substitution. In each case, N-SH2T42A is more sensitive to the peptide mutation than N-SH2WT. (G) Heatmaps of N-SH2WT and N-SH2T42A for PD-1 pTyr 223 (ITIM) scanning mutagenesis screen (average of 5 replicates). The wild-type residue on each position is indicated by a black square. Blue indicates that the mutant binds worse than the wild-type ITIM, white indicates no effect on binding relative to wild-type, and red indicates that the mutant binds better than the wild-type ITIM. Source data for the heatmaps in panel (G) can be found in Table S4.