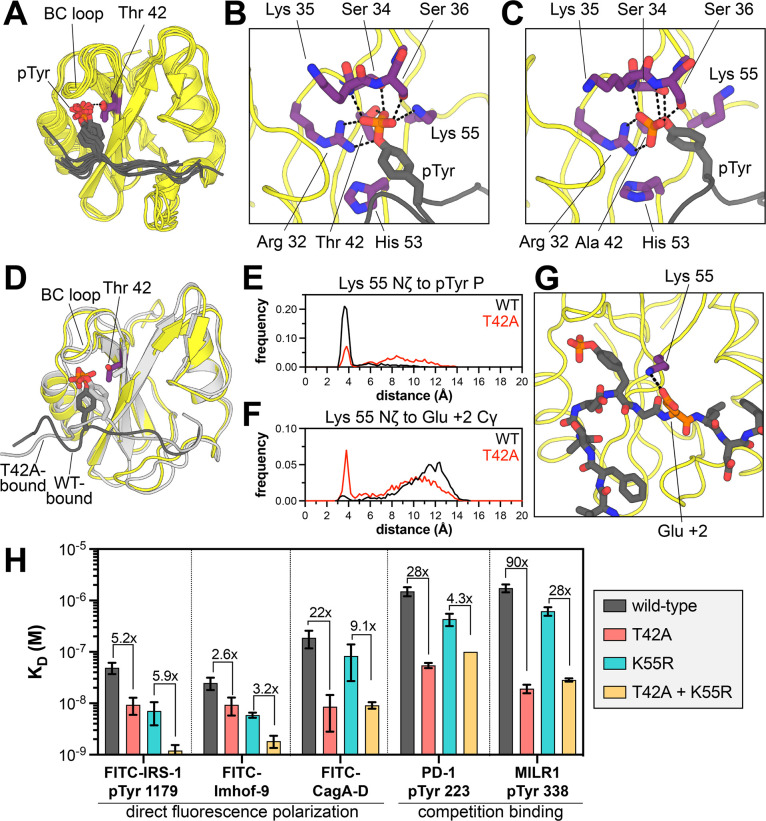
Figure 5. Structural impact of the T42A mutation on phosphotyrosine and proximal sequence recognition.
(A) Hydrogen bonding of Thr 42 in SHP2 N-SH2WT to the phosphoryl group of phosphopeptide ligands, as seen in several crystal structures (PDB codes: 6ROY, 1AYA, 1AYB, 3TL0, 5DF6, 5X94, and 5X7B). (B) Structure of N-SH2WT bound to the PD-1 pTyr 223 (ITIM) peptide at the end of a 1 μs MD simulation, highlighting a hydrogen bond network and other key interactions around the phosphotyrosine residue. (C) Structure of N-SH2T42A bound to the PD-1 pTyr 223 (ITIM) peptide at the end of a 1 μs MD simulation, highlighting a distinct hydrogen bond network around the phosphotyrosine residues, relative to that seen for N-SH2WT. (D) Overlay of the states shown in panels B and C, highlighting a change in position for the phosphotyrosine residue and peptide main chain upon T42A mutation. The N-SH2WT state is in yellow with a dark-gray ligand. The N-SH2T42A state is in light gray, with a light gray ligand. (E) Distribution of distances between the Lys 55 Nζ atom and the phosphotyrosine phosphorus atοm in simulations of the PD-1 pTyr 223 peptide bound to N-SH2WT (black) or N-SH2T42A (red). (F) Distribution of distances between the Lys 55 Nζ atom and the +2 Glu Cδ atom in simulations of the PD-1 pTyr 223 peptide bound to N-SH2WT (black) or N-SH2T42A (red). (G) An ion pair between Lys 55 and the +2 Glu residue (Glu 225) in the PD-1 pTyr 223 (ITIM) peptide, frequently observed in N-SH2T42A simulations. (H) Effects of the T42A mutation in the context of the K55R mutation. The enhancement in binding affinity by the T42A mutation is attenuated by the K55R mutation for some peptides (CagA-D, PD-1 pTyr 223, and MILR1 pTyr 338) but not others (IRS1 pTyr 1179 and Imhof-9).