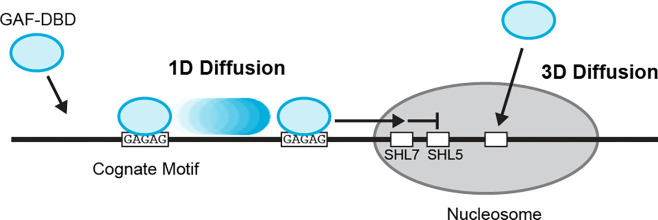
Figure 5.
Model for GAF-DBD target search on chromatin. GAF-DBD uses two search modes to locate its target on chromatin. In the 1D sliding mode, GAF-DBD lands on an off-target location on free DNA, then slides on DNA to locate the adjacent GAF motif (“GAGAG”). It can escape the cognate site to locate another cognate site nearby. 1D search allows GAF-DBD to invade into the nucleosome edge and locate cognate motifs at SHL7. Alternatively, GAF-DBD can also directly associate with a cognate motif in the nucleosome core from 3D space. This 3D search mode allows GAF to effectively target nucleosomal motifs that are inaccessible by 1D sliding.