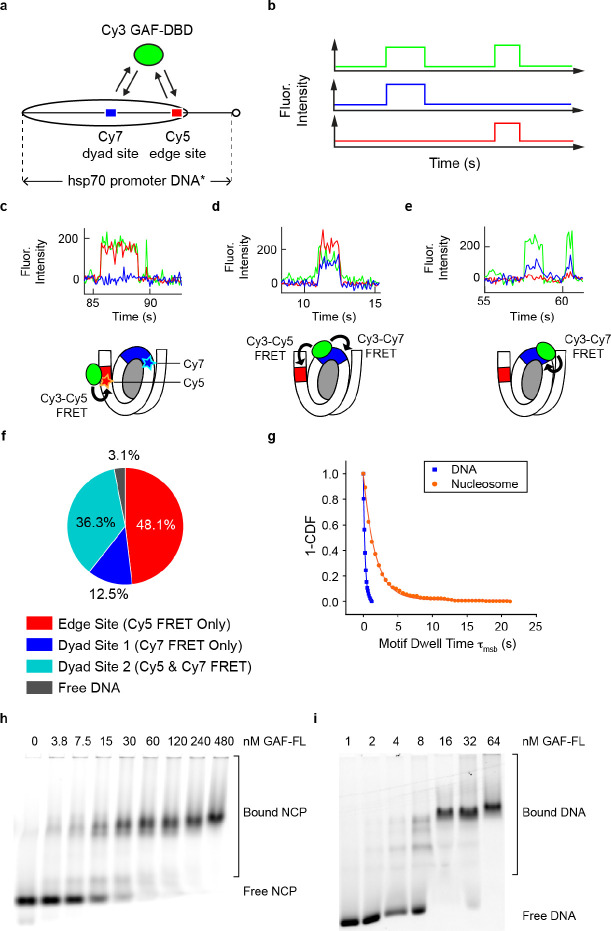
Figure 4.
3D diffusion dominates for inner nucleosomal targets. a, Drosophila hsp70 promoter nucleosome construct (0-N-40) for investigating GAF-DBD search when two binding sites are within nucleosome core. b, Idealized single-molecule trajectories for GAF-DBD locating two nearby cognate sites. c-e, Representative single-molecule trajectories on this nucleosome construct. 0.4 nM Cy3-GAF-DBD was used for these experiments. f, Binding event categories on the nucleosome. g, 1-CDF of motif dwell times for nucleosome (orange) compared with free DNA (blue). Free DNA data are from Cy7-Site 1 dwell time (Fig. 2e, 6.25 mM MgCl2). h, EMSA showing GAF-FL binding to Cy5-NCP formed on hsp70 promoter DNA fragment. i, EMSA showing GAF-FL binding to Cy5-DNA. The same DNA was used to reconstitute Cy5-NCP in h.