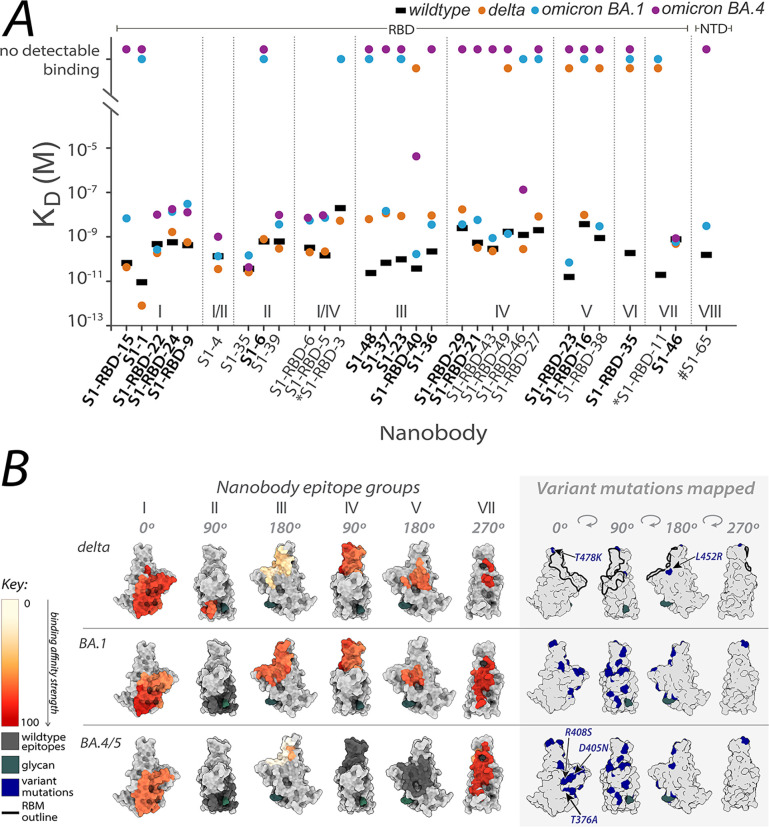
Figure 2. Affinities of the nanobody repertoire against SARS-CoV-2 variants.
(A) Each nanobody is plotted against their affinity (KD) for SARS-CoV-2 Spike S1 from wild-type, delta and omicron BA.1 and BA.4 strains. Nanobodies are characterized into their respective epitope groups according to Mast et al (Mast, Fridy et al. 2021). (B) The structure of the RBD of spike delta (PDB ID: 7SBO), omicron BA.1 (PDB ID: 7T9K) and omicron BA.4 RBD (modeled) is shown, with heat-mapped epitopes of binding colored from pale white (epitopes with weak binding against SARS-CoV-2) to dark red (strong binding against SARS-CoV-2). The nanobodies that contributed to epitope mapping are in bold in panel A. The color bar scale for each epitope shows the binding affinity strength of each nanobody epitope, which is the normalized −log ratio of nanobody binding (KD) of variant to wild-type SARS-CoV-2 Spike S1. A higher value of −log ratio corresponds to strong binding of the nanobody to the variant. S1-RBD-16 showed binding to omicron BA.1 in ELISA. *S1-RBD-11 was not tested against omicron BA.4 and #S1–65 was not tested against BA.1. All structure representations were created on ChimeraX (Pettersen, Goddard et al. 2021).