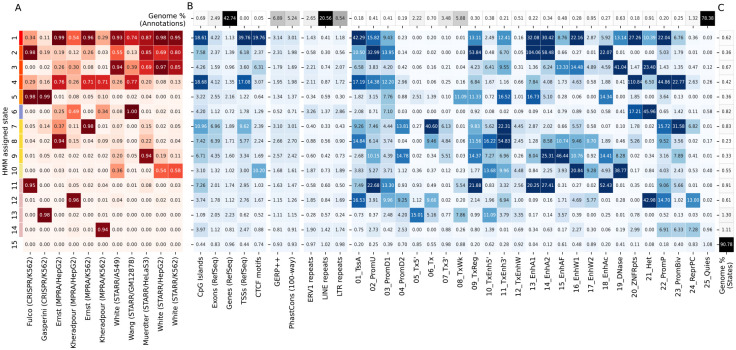
Figure 3: ChromScoreHMM emission parameters and enrichments.
(A) Emission parameters of a ChromScoreHMM model learned based on combinatorial and spatial patterns of top scoring predictions of each expert (top 2% of predictions, Methods). Each row of the heatmap corresponds to a ChromScoreHMM state (states 1–15, color legend on left margin) and each column a different input expert model. Emission parameter values correspond to the probability in that state of observing a top scoring prediction for that expert model. (B) Overlap fold enrichments for (1) sequence and gene annotations: CpG islands [40], exons, gene bodies and transcription start sites from RefSeq [41], CTCF motifs from HOMER [42], (2) evolutionary conservation related annotations: GERP++ [43], PhastCons 100 vertebrates conservation [44], (3) ERV1, LINE and LTR repeat elements from RepeatMasker [45], (4) ChromHMM annotations, 25-state model [13, 39]. Top row: Percentage of the genome occupied by the annotation. (C) Percentage of the genome assigned to each ChromScoreHMM state. See Supplementary Figures S7 and S8 for additional enrichments. Red shading: emission parameters, blue shading: fold enrichments, black shading: genome percentages. Enrichments and percentages are medians across cell types.