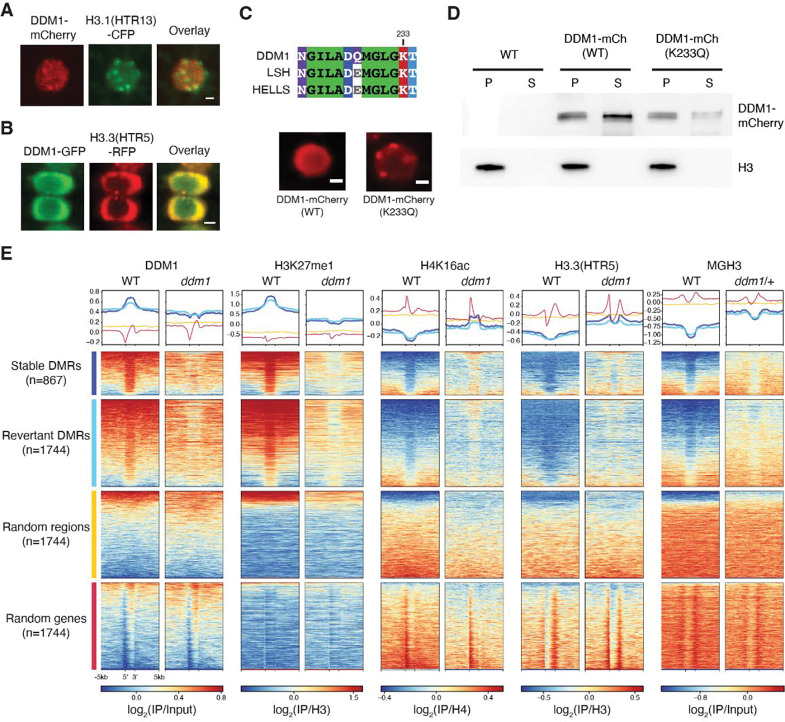
Figure 6. DDM1 remodels H3.3 and H3.1 in interphase at differentially methylated targets of DDM1.
(A) Subnuclear localization of DDM1-mCherry in root tip cells as compared to H3.1(HTR13)-CFP during presumptive late S-phase (marked by H3.1-labeled chromocenters). (B) Subnuclear localization of DDM1-GFP as compared to H3.3(HTR5-RFP) during interphase. The scale bar indicates 2 μm. See also Supplemental Videos S1 and S2. (C) Conserved amino acids of the ATP binding sites for DDM1 and orthologs, LSH (mouse) and HELLS (human). K233Q Walker A mutation disrupts ATP binding and causes enhanced chromocenter localization of DDM1-mCherry fusion in a WT background. Scale bar indicates 2 μm. (D) Western blot of DDM1-mCherry (DDM1-mCh) for the wild-type (WT) and mutant form (K233Q) from soluble (S) and chromatin/pellet (P) fractions indicates failure to release catalytic mutant from chromatin. H3 was used as loading control. Non-transgenic WT was used as negative control. (E) Comparisons of DDM1, H3K27me1, H4K16ac and H3.3(HTR5) chromatin association by ChIP-seq in WT and ddm1 leaves, as well as MGH3 in pollen from WT and ddm1/+ plants. Stable and revertant differentially methylated regions (DMRs) lost DNA methylation in ddm1 mutants, and stable DMRs never regained methylation when DDM1 was reintroduced 74. Thus, DMRs represent epigenetic targets of DDM1. Heatmaps and metaplots were generated using DeepTools 95, where each region was scaled to 2kb with 5kb upstream and 5kb downstream with a binsize of 10bp, and sorted based on DDM1 levels in WT. Metaplots above each heatmap show the mean value for each region. Random regions are revertant DMRs reshuffled randomly in the genome, whereas random genes correspond to the same number of protein coding genes selected at random (see Table S2). DDM1 and H3.1 (H3K27me1) are specifically enriched at DDM1 targets (DMRs), while H4K16Ac, H3.3 and MGH3 are specifically depleted, except in ddm1 mutants. Similar analysis was performed on all transposable elements for comparison (Figure S5).