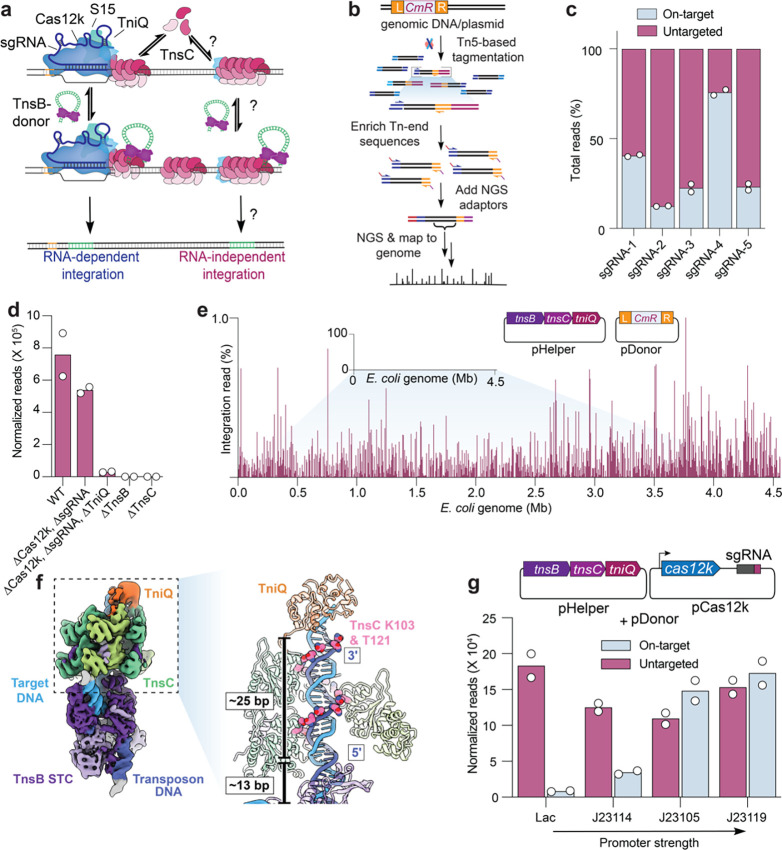
Fig. 1 |. Type V-K CASTs direct frequent Cas12k- and RNA-independent transposition events.
a, Schematic of type V-K CAST transposition occurring at on-target sites (RNA-dependent) and untargeted sites (RNA-independent). b, Experimental pipeline used for tagmentation-based transposon insertion sequencing (TagTn-seq) for in vitro and genomic samples. c, Fraction of total genome-mapping integration reads detected at on-target and untargeted sites for the wildtype pHelper expression plasmid across multiple guides (sgRNA-1 to sgRNA-5). d, Total genome-mapping reads detected for WT pHelper or pHelper with the indicated deletions, normalized and scaled (Methods). e, Zoomed-in view of integration reads comprising 1% or less of E. coli genome-mapping reads, in an experiment performed without the Cas12k and guide RNA. f, Cryo-EM reconstruction of the untargeted transpososome reveals the assembly of TniQ (orange), TnsC (green), and TnsB (purple) in a strand-transfer complex (STC). The target DNA and transposon DNA are represented in light blue and dark blue, respectively. For visualization, a composite map was generated using two local-resolution filtered reconstructions from the focused refinements (Methods). Zoomed-in and cutaway view showing TnsC forming a helical assembly on the target DNA, positioning residues K103 and T121 (pink) adjacent to one strand of the target DNA (dark blue). 5’ and 3’ ends of the TnsC-interacting DNA strand are indicated. Two turns of TnsC and TnsB footprint on DNA until TSD cover approximately 25 and 13 base pairs (bp), respectively. Only selected TnsC monomers are represented in the cutaway for clarity. g, Cas12k and the sgRNA were cloned onto a separate vector, and the promoter driving Cas12k expression was varied. Reads detected at on-target and untargeted sites during transposition assays were normalized and scaled (Methods). For c, d, e, and g, the mean is shown from n = 2 independent biological replicates.