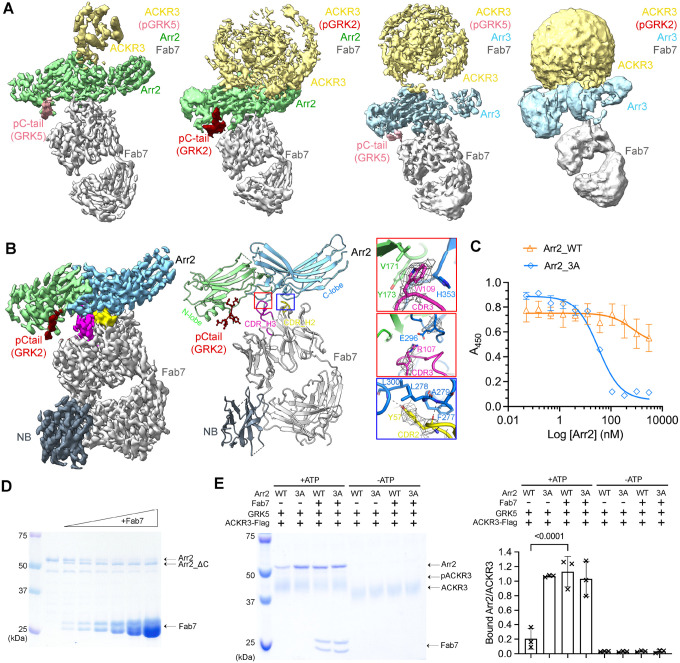
Figure 2. Structural and functional characterization of Fab7, a new arrestin conformational sensor.
(A) Sharpened maps of the 3.2 Å pACKR3(GRK5)–Arr2–Fab7, the 3.5 Å pACKR3(GRK2)–Arr2–Fab7, the 3.9 Å pACKR3(GRK5)–Arr3–Fab7, and the 7.3 Å pACKR3(GRK2)–Arr3–Fab7 complexes. ACKR3 in all four complexes is solubilized in LMNG/CHS detergent micelles. (B) Sharpened map and model of the 3.0 Å pACKR3(GRK2)–Arr2–Fab7–NB in POPC/POPS nanodiscs (PDB entry XXXX). The density of the ACKR3 TM domain and the nanodisc is not evident. Insets show interfacial details discussed in the text. (C) ELISA analysis of Fab7 competition assay reveals that preactivated Arr2 (IC50 ~35 nM) competes for Fab7 binding more efficiently than Arr2 WT (IC50 > 5 μM). Error bars represent S.D. from three technical replicates. (D) Limited trypsin digestion of Arr2 WT in the presence of increasing concentrations of Fab7. (E) A Flag pulldown assay shows that Fab7 significantly promotes Arr2 WT binding to pACKR3(GRK5), but it does not increase Arr2 WT or 3A binding non-phosphorylated ACKR3. One representative gel is shown. The ratios between the density of bound Arr2 and that of bound ACKR3 are compared using one-way ANOVA followed by a Dunnett’s multiple comparison test (P < 0.0001). Error bars represent S.D. from three technical replicates.