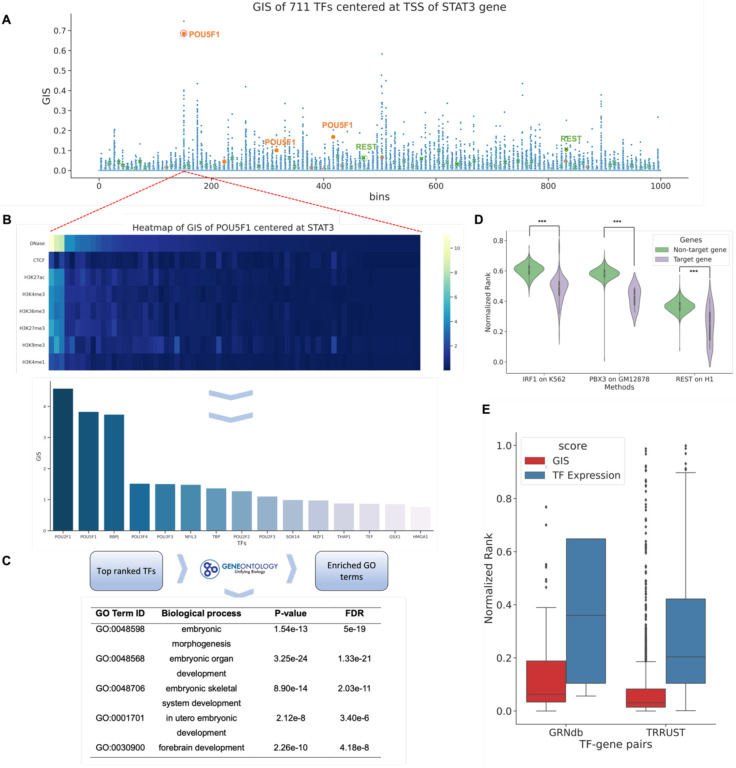
Fig. 5. Gradient importance scores (GIS) uncover regulatory transcription factors.
(A) Genomic regions around TSS of the STAT3 gene and expression data on ESC were fed into EpiGePT. The scatter plot represents the GIS scores of core TFs on each genomic bin. Each dot represents the GIS score of a core TF on a specific genomic bin. In specific bins, key TFs in ESC, such as NR5A2 and POU1F5 highlighted in the figure, exhibit high ranks in the GIS scores. (B) Heatmap of GIS for 10% important TFs surrounding STAT3 gene, specifically focusing on bins with POU1F5 ranked 3rd. Each row represents a predicted epigenomic signal, and the TFs are sorted based on their GIS on DNase signals. Bar plot of the top 15 TFs with the highest GIS scores. (C) Based on the top 10% ranked TFs mentioned above, gene ontology enrichment analysis revealed significant enrichment in biological processes related to embryonic development and cellular differentiation. (D) Based on TF ChIP-seq data, all 23,635 human genes were classified into target genes and non-target genes. The results revealed that TFs exhibited significantly higher ranks on potential target genes compared to non-target genes. (E) The distribution of the rank of TFs in the GIS and expression value among the 2,705 TF-gene pairs from the TRRUST database and 1,066 TF-gene pairs derived from genotype-tissue expression (GTEx) data of the liver sourced from the GRNdb database. The analysis reveals that the median rank of TFs from the TRRUST database is significantly lower than 0.06 (one-side p-value < 3.12 e-18) and the median rank of TFs obtained from the GRNdb database is significantly lower than 0.5 (one-side p-value < 2.50 e-140).