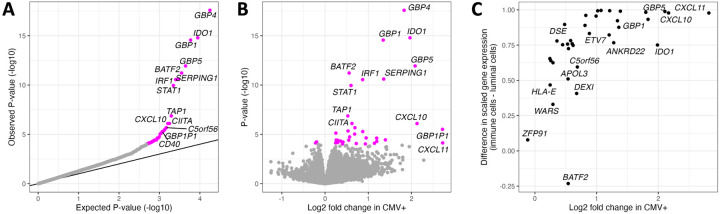
Figure 2.
Differential gene expression analysis comparing CMV− to CMV+ milk samples. (A) QQ-plot from the results of differential gene expression analysis. The x-axis plots the expected P-value for the number of genes tested following a uniform distribution of P-values from 0 to 1, and the y-axis plots the observed P-values. Genes whose P-value was below the false discovery rate threshold of 5% are colored in magenta. (B) Volcano plot comparing estimated effect sizes of CMV+ on milk gene expression (x-axis) with each gene’s P-value (y-axis). Genes whose P-value was below the false discovery rate threshold of 5% are colored in magenta. (C) Comparison of log fold change in CMV+ samples from our bulk RNA-seq data (x-axis) vs. gene expression in a publicly available human milk single cell RNA-seq dataset36 (y-axis). Gene expression from milk single cells is plotted as the difference between scaled gene expression in immune cells and mammary luminal cells, to display that genes more highly expressed in our CMV+ milk samples tended to be more highly expressed in the immune cells in milk.