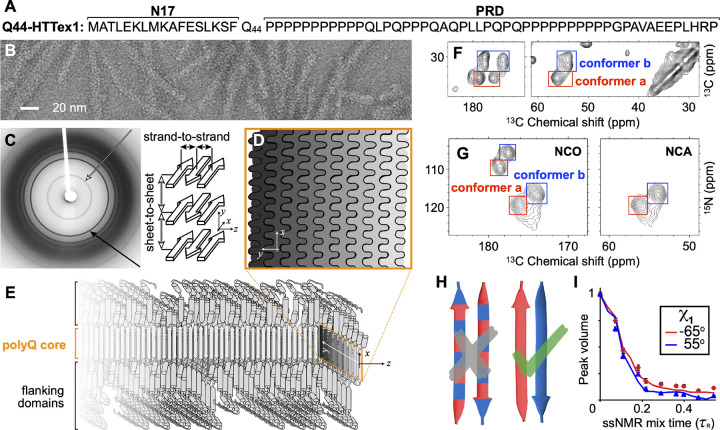
Figure 1. Select structural data on HTT Exon 1 aggregates.
(A) Sequence of Q44-HTTex1. N- and C-terminal flanking domains marked as N17 and PRD, respectively. (B) Negatively-stained TEM of Q44-HTTex1 aggregates formed in vitro; average fiber width ~6.5 nm. (C) X-ray fiber diffraction of K2Q31K2 fibrils, detecting the cross-β reflections of the amyloid core that represent the inter-strand and inter-sheet distances shown on the right. (D) Cross-section of the (6–7-nm wide) block-like core showing the layering and interdigitation of (differently shaded) β-sheets. (E) Schematic model of a Q44-HTTex1 fibril, showing the N17 and PRD flanking domains outside the polyQ core. (F) 2D 13C–13C ssNMR spectrum of 13C,15N-labeled Q44-HTTex1 fibrils showing backbone and sidechain cross-peaks of the dominant signals of the polyQ fibril core. Peaks for Cα–CX correlations of two distinct “a” and “b” Gln conformers are marked with red and blue boxes, respectively. (G) 2D NCO and NCA ssNMR spectra of Q44-HTTex1 fibrils, with the conformer “a” and “b” again marked. (H) Pairs of antiparallel β-strands, schematically showing two previously proposed arrangements of the “a” (red) and “b” (blue) types of Gln. Given the ssNMR data (panels F–G), only the right one fits the experimental data. See also Supplementary Fig. S1 (I) ssNMR measurements sensitive of the side-chain dihedral angle χ1, with simulated curves for “a” (red) and “b” (blue) types of Gln. Panels B–C and I adapted with permission from Ref. 8, D–E from 38, and G from 39.