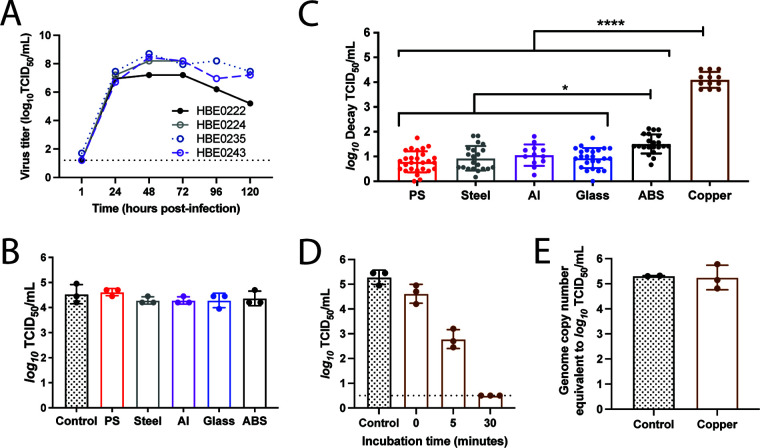
FIG 1.
The stability of H1N1pdm09 is dependent on surface material at low relative humidity. (A) Propagation of H1N1pdm09 in four different HBE patient cultures. Each culture can be related to a different pathological state: culture 235 (nonpathological), culture 222 (chronic obstructive pulmonary disease), culture 243 (nonpathological), and culture 224 (idiopathic pulmonary fibrosis). HBE cultures were infected with 103 TCID50 of virus per well. Virus was collected at the indicated times and titrated on MDCK cells. Virus samples from 24 to 96 h postinfection for each HBE patient culture were then pooled for use in the stability experiments. (B) Ten 1-μL droplets of H1N1pdm09 were deposited on the indicated surfaces and immediately recovered. Virus titrated as for panel A. Each dot represents individual replicates for each HBE culture. (C) Ten 1-μL droplets of H1N1pdm09 (A/CA/07/2009) were incubated at 23% RH on the surface of the indicated materials for 2 h. Recovered virus was titrated by TCID50 assay, and viral decay was expressed as the loss of infectivity compared to a control (10 μL of virus in a sealed Eppendorf tube outside the chamber) that was incubated for the same amount of time. Each data point represents a replicate of H1N1pdm09 propagated from three to four different HBE cultures. Data are means and standard deviations. Asterisks indicate significantly different mean levels of decay between indicated surface materials, as determined by one-way ANOVA with Tukey’s multiple-comparison test (*, P < 0.05; ****, P < 0.0001). No infectious virus was detected after incubation on copper, and the log10 decay values (TCID50 per milliliter) were maximal in all replicates. (D) Ten 1-μL droplets of H1N1pdm09 were deposited onto copper. The virus was recovered from the copper surface either immediately after droplet deposition (0 min) or after incubation for 5 or 30 min at room RH, which ranged between 41 and 43%. Virus was titrated by TCID50 assay, and data are means and standard deviations. The dashed line indicates the limit of detection. Data shown are representative of three independent experiments in different HBE cultures. (E) Ten 1-μL droplets were recovered following a 2-h incubation on copper at 23% RH. Quantitative PCR for the influenza A virus M gene was used to determine the amount of viral RNA in each sample compared to a known quantity of viral RNA. Each data point in panels C to E represents an individual replicate within a study.