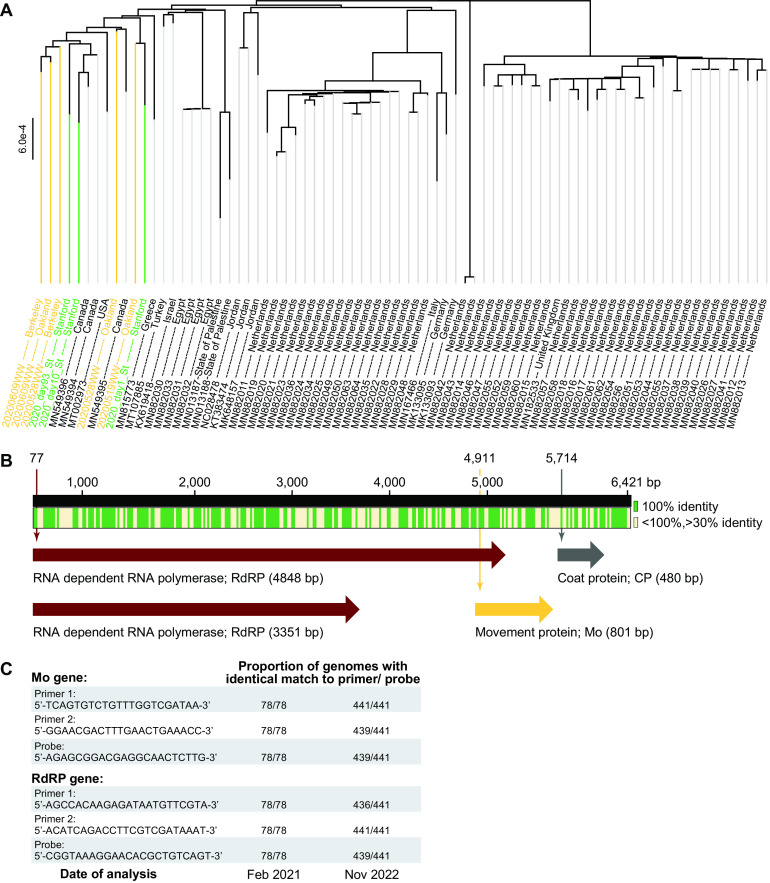
FIG 2.
Analysis of newly assembled ToBRFV genomes and generation of primer/probe sets for ddRT-PCR. (A) Phylogenetic tree of 78 nearly complete genomes of ToBRFV, including eight genomes generated in the current study from wastewater and stool. All genomes are listed by their NCBI accession number and source location. Seventy preexisting genomes are listed in black font, five genomes derived from wastewater samples are in yellow, and three from stool samples are in green. (B) Summary of multiple-sequence alignment and gene annotation across the 78 ToBRFV genomes. Green indicates regions that are 100.0% conserved across all genomes, while cream marks those that are greater than 30.0% but less than 100.0% conserved. Two variants of the RdRP-encoding gene are found at 77 bp and are either 4,848 bp or 3,351 bp in size. The Mo protein-encoding gene is found at 4,911 bp and is 801 bp in size. The CP-encoding gene is found at 5,714 bp and is 480 bp in size. Genomic locations are based on the genome ID NC_028478. (C) Sequences of primer/probe sets generated in the current study, in February 2021, aimed at targeting the Mo and RdRP genes across all known genomes. Since the number of known genomes grew from February 2021 to November 2022, the final column indicates the proportion of the 441 current genomes bearing sequences identical to the designed primer/probe sets.