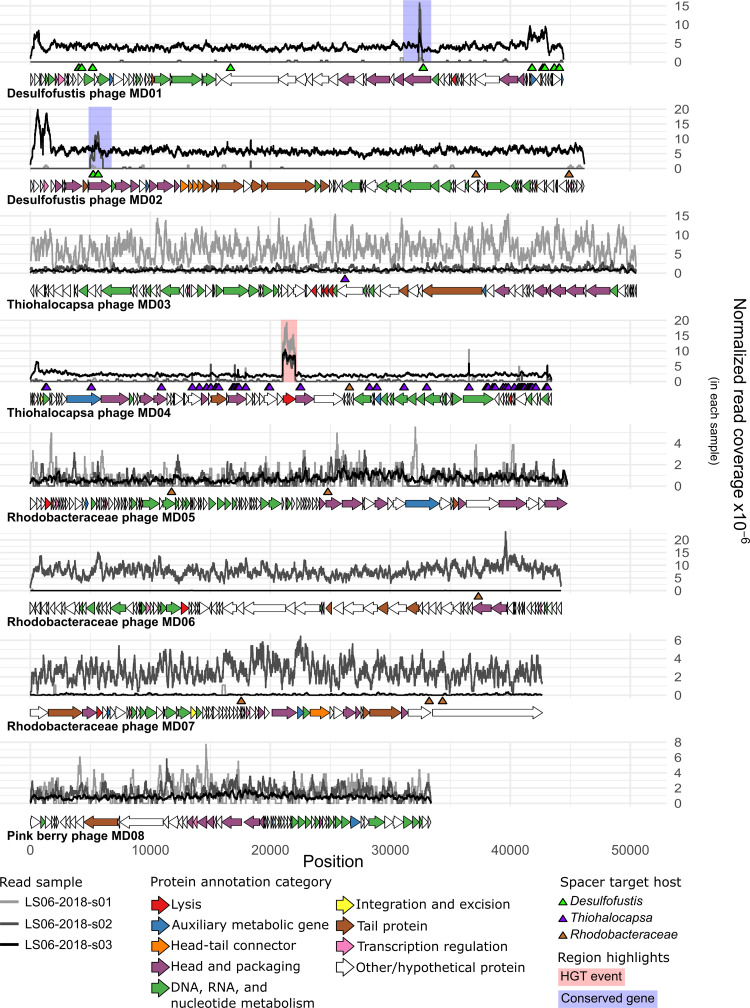
FIG 2.
Complete phage genomes vary in abundance across samples and are targeted by bacterial CRISPR spacers. Normalized read coverage by position for each sample are given. Coverage values were normalized to the total number of trimmed and filtered reads for each sample. The horizontal arrows indicate the ORFs predicted by PHANOTATE (54), and their colors correspond to their predicted functional categories. The triangles indicate the genome positions of protospacers, and they are colored according to the host taxonomy of the corresponding spacer. Regions highlighted with a blue background indicate a conserved gene between phage genomes, as inferred by Clinker (62). Regions highlighted with a red background were found to be an HGT event between the phage and the host.