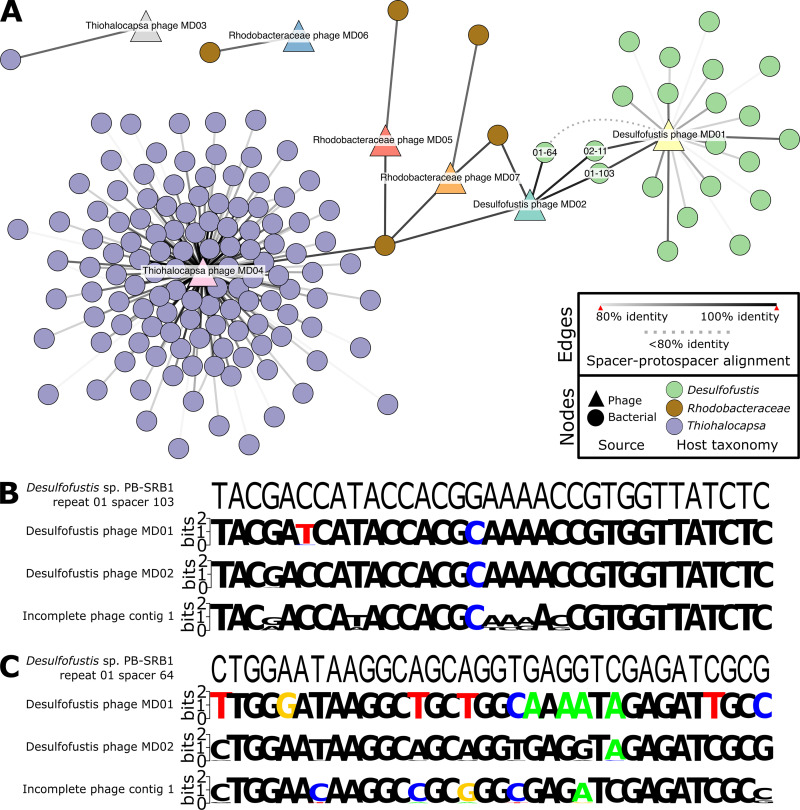
FIG 5.
CRISPR spacer-phage genome alignments reveal hosts and a conserved protospacer. (A) Spacerblast (71) alignment results with at least 80% nucleotide identity over the entire spacer length were visualized in Cytoscape v3.9.0 (78). The circular nodes represent unique spacers from bacterial contigs and are colored by taxonomy. The triangular nodes are phage contigs. Solid edges represent the percent nucleotide identity over the entire spacer length. The dashed edge shows the connection between Desulfofustis phage MD01 and Desulfofustis sp. PB-SRB1 repeat 01 spacer 64, which is below 80% identity and included in panel C. The nucleotide sequences of phage protospacers within conserved capsid genes that were identified on phage contigs were aligned to (B) Desulfofustis sp. PB-SRB1 repeat 01 spacer 103 and (C) Desulfofustis sp. PB-SRB1 repeat 01 spacer 64. Spacer 02-11 is the reverse complement of 01-103 and is not shown. The resulting metagenome-wide variation in protospacer sequences from mapping reads to protospacers is shown as sequence logos (73, 79).