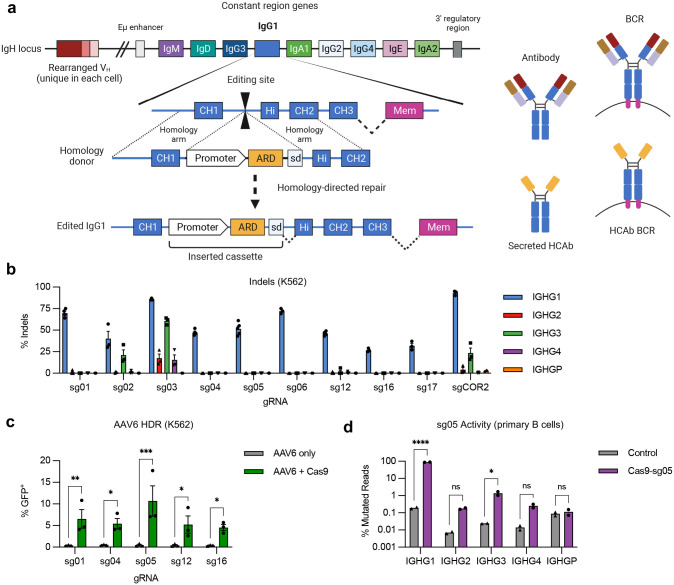
Figure 1. Genome editing at the constant region of the IgH locus.
(a) Antibody H chains are encoded by a rearranged variable domain (VH), spliced to a constant region that can be altered by class switch recombination. Alternate splicing within the constant region generates secreted antibody or membrane anchored (BCR) isoforms. Genome editing to insert custom antigen recognition domains (ARD) downstream of CH1 exons in the IgH constant regions can create Heavy chain only antibodies (HCAbs). The example shown is targeting human IgG1, with the expanded view showing the constant region exons. Genome editing is directed by homology donors containing 500–750 bp homology arms, flanking CRISPR/Cas9 target sites in the intron downstream of CH1. Homology directed repair of the targeted DNA break inserts a cassette comprising a B cell-specific promoter, a custom ARD, and a splice donor to direct splicing to downstream endogenous exons. (b) K562 cells were edited with Cas9 RNPs programmed by gRNAs targeting the IgG1 CH1 intron and indels measured at on-target (IGHG1) and off target (IGHG2-P) genes by Sanger sequencing and ICE (n = 3). (c) K562 cells were edited by Cas9 RNPs plus AAV6 homology donors containing a GFP expression cassette, matched for each gRNA. GFP expression was measured after 3 weeks, to dilute out episomal AAV genomes (n = 3). (d) On- and off-target activity of sg05 was measured at indicated IGHG genes in primary human B cells (n = 2), 5 days after editing with sg05 RNPs, by targeted amplicon deep sequencing, with percentage mutated reads calculated as insertions, deletions, ≥ 2 bp changed. See also Extended Data Fig. 1d,e. Error bars show mean ± SEM. Statistical comparisons (c-d) were performed by 2-way ANOVA. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.