Figure 1.
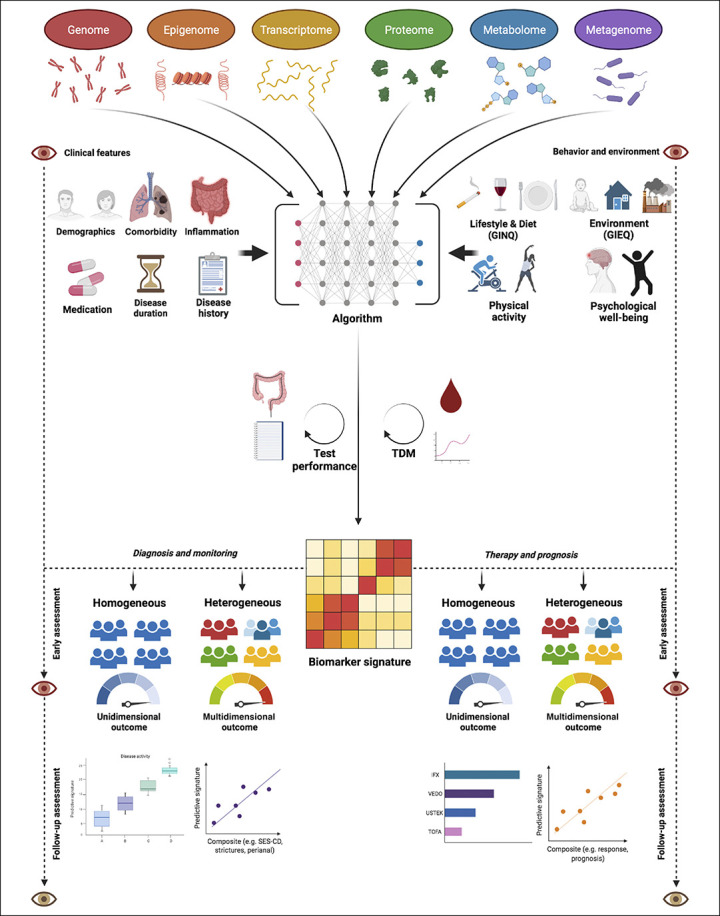
Multi-omics-driven biomarker discovery as strategy to develop personalized medicine for patients with IBD. An unbiased, data-driven approach may reveal key pathophysiological signatures while necessitating the integration of “phenome” data (i.e. clinical [upper left] and environmental [upper right] features) in order to improve clinical outcomes of patients with IBD. Artificial intelligence algorithms may help to allow the generation of individualized biomarker signatures from this molecular data-driven approach. After successful integration and validation, such individualized predictive biomarker signatures may aid in diagnosis and monitoring [lower left] as well as predicting therapeutic success and disease prognosis [lower right]. Depending on the disease outcome, different approaches could be followed to clinically integrate these signatures. One approach entails studies of patients with IBD having heterogeneous features that may be suitable for multi-dimensional (disease modification) outcome assessment, e.g. composite outcomes of disease activity or therapeutic response. Another approach consists of studies focusing on patients with IBD having homogeneous features to allow the assessment of the true objectivity of a certain multi-omics-derived biomarker by matching a specific group of patients to its most suitable outcome measure. Both approaches, however, require ‛quality control': for example, in case of diagnostic signatures, it is crucial that the gold standard outcome is objectively and accurately determined, whereas in case of therapeutic signatures, it is crucial that drug exposure is sufficiently guaranteed (e.g. by TDM). Eventually, developed signatures need long-term follow-up to assess the durability of their effectiveness in predicting disease outcomes. Abbreviations: GIEQ, Groningen IBD Environmental Questionnaire (24); GINQ, Groningen IBD Nutritional Questionnaire (25); TDM, therapeutic drug monitoring.
