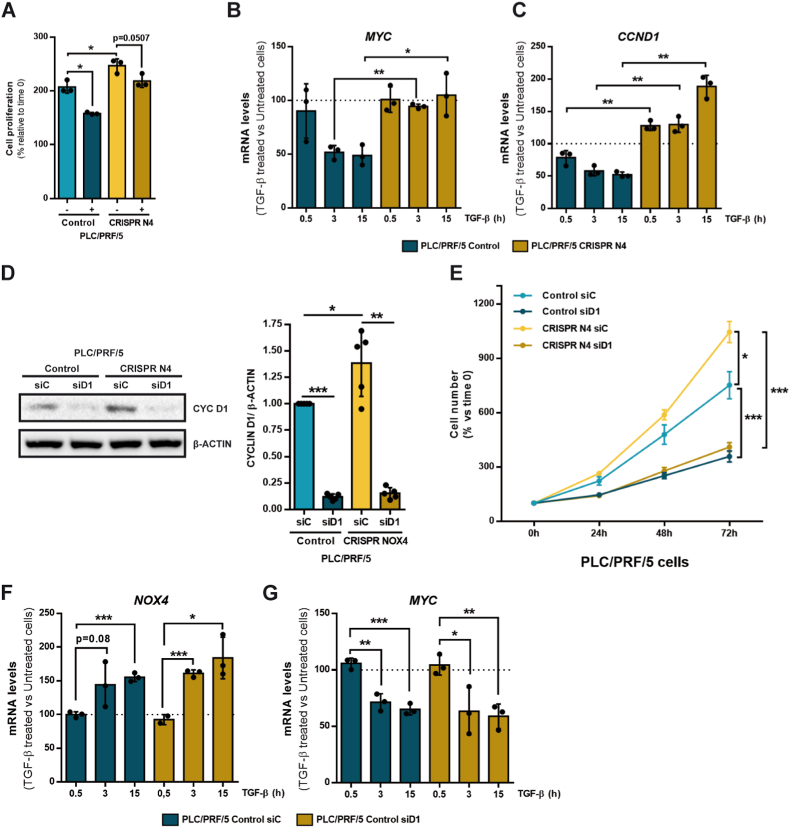
Fig. 2.
Role of NOX4 in the response to TGF-β in terms of cell proliferation. Analysis made in PLC/PRF/5 (Control and CRISPR NOX4) cells. A) Cell viability assay analyzed by crystal violet staining after 72h of TGF-β treatment, normalized to initial time (time 0h). Data represents mean ± SD of triplicates from one representative experiment. B) Relative MYC and C)CCND1 mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, after TGF-β treatment at 0, 0.5, 3 and 15 h. Represented as percentage of TGF-β treated versus untreated cells in each time. D) Cyclin D1 protein levels analyzed by Western blot after transiently transfecting either with control or Cyclin D1 specific siRNA sequences. β-Actin was used as loading control. Representative experiment (left) and densitometric analysis of Cyclin D1 levels relative to β-Actin (right). E) Cell proliferation assay analyzed by crystal violet staining after transiently transfecting with either control or Cyclin D1 specific siRNA sequences, normalized to initial time (time 0h). F) Relative NOX4 and G)MYC mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, in Control CRISPR PLC/PRF/5 cells transiently transfecting with either control or Cyclin D1 specific siRNA sequences: effect of TGF-β treatment at 0, 0.5, 3 and 15 h. Data are represented as percentage of TGF-β treated versus untreated cells in each time. In all the panels, data are Mean ± SD (n = 3). *p<0.05 **p<0.01 ***p<0.001. (siC: Control siRNA; siD1: Cyclin D1 siRNA). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)