Fig. 6.
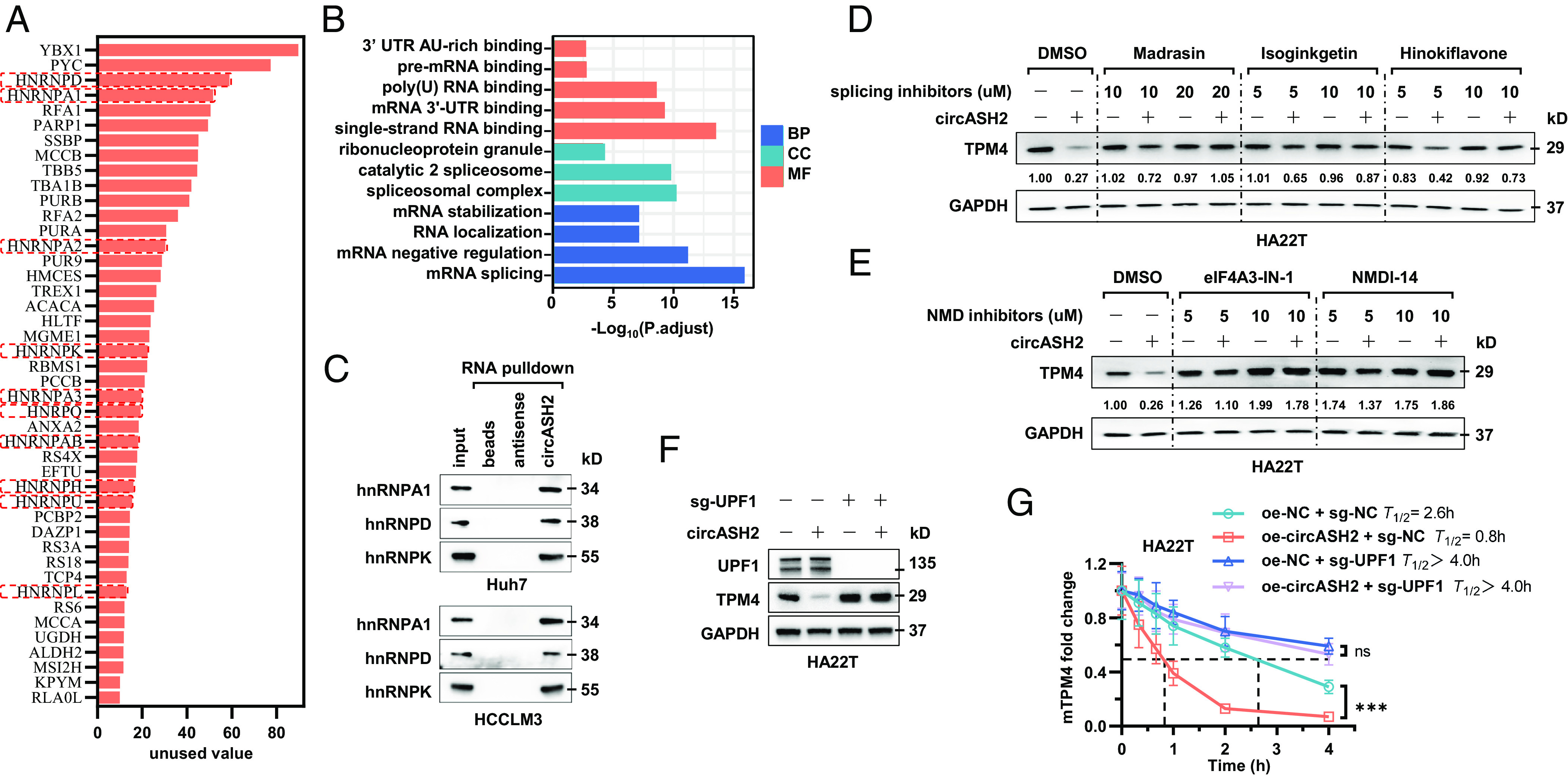
circASH2/hnRNP complex accelerates mRNA degradation via nonsense-mediated decay. (A) A diagram showing potential circASH2-binding proteins identified by MS (listed in the order of unused value). hnRNPs are highlighted with red boxes. (B) Gene ontology (GO) analysis of circASH2-interacting proteins, including molecular function (MF), biological process (BP), and cellular component (CC). (C) Three representative hnRNPs (hnRNPA1, hnRNPD, and hnRNPK) were confirmed as circASH2-binding proteins by immunoblot analysis in Huh7 and HCCLM3 cells. (D) HA22T cells overexpressing circASH2 or control vector were treated with Dimethylsulfoxide (DMSO), Madrasin (10 to 20 μM), isoginkgetin (5 to 10 μM), or hinokiflavone (5 to 10 μM) for 48 h. The endogenous TPM4 level was probed, while GAPDH was used as an internal reference. (E) HA22T cells overexpressing circASH2 or control vector were treated with DMSO, eIF4A3-IN-1 (5 to 10 μM), NMDl-14 (5 to 10 μM) for 48 h. The endogenous TPM4 level was probed, while GAPDH was used as an internal reference. (F) UPF1 was depleted in HA22T cells, and circASH2-induced downregulation of endogenous TPM4 was assessed by immunoblot analysis. GAPDH was used as an internal reference. (G) RT-qPCR detection of TPM4 mRNA at the indicated times in indicated HA22T cells treated with 2 μg/mL actinomycin D. The dashed line shows the half-life of TPM4 mRNA (mean ± SD, ***P < 0.001 and n.s., not significant, two-way ANOVA test).
