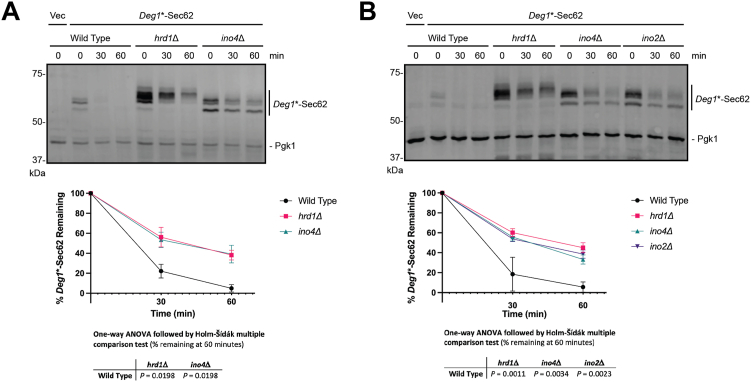
Figure 2.
Ino2 and Ino4 are required for Deg1∗-Sec62 degradation.A, WT yeast or yeast lacking either HRD1 or INO4 were transformed with a plasmid encoding Deg1∗-Sec62 or an empty vector and subjected to cycloheximide chase and western blot analysis to detect Deg1∗-Sec62 and Pgk1. B, as in (A), but with WT yeast or yeast lacking either HRD1, INO4, or INO2. Means of percent Deg1∗-Sec62 remaining for three to four biological replicates are plotted. Error bars represent the SEM. Means of percent Deg1∗-Sec62 remaining at 60 min were evaluated by one-way ANOVA followed by Holm-Šídák multiple comparison tests (only pairs relative to WT yeast were compared).