Figure 1. The flavonoid biosynthetic pathway.
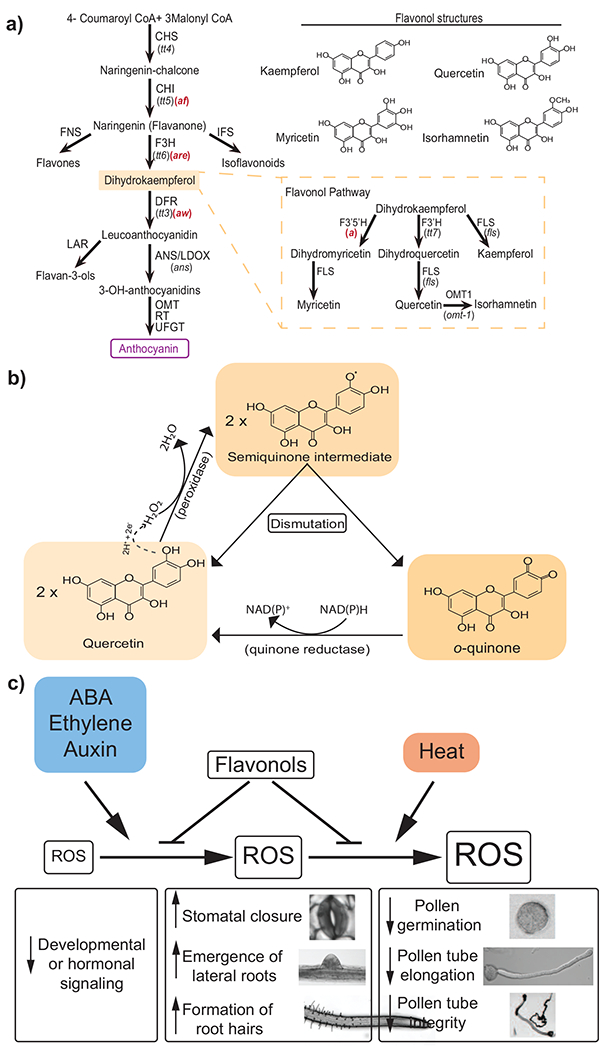
Enzymes are indicated next to arrows with mutant names in parentheses under the enzyme abbreviation. Arabidopsis mutants are in black and tomato mutants are in red. The flavonol biosynthesis branch is illustrated in the yellow dashed box. Chemical structure of specific flavonols are presented on the upper right. Abbreviations: CHS, chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; F3H, flavanone 3-hydroxylase; IFS, isoflavone synthase; DFR, dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase; ANS/LDOX, anthocyanidin synthase/leucoanthocyanidin dioxygenase; RT, rhamnosyl transferase; UFGT, UDP-glucose flavonoid 3-O-glucotransferase; F3’5’H, flavonoid 3’5’-hydroxylase; F3’H, flavonoid 3’-hydroxylase; FLS, flavonol synthase; OMT-1, O-methyltransferase-1. Figure adapted from [64 & 66], B. The predicted mechanism by which the flavonol quercetin donates electrons to convert hydrogen peroxide to water is shown, along with a potential mechanism by which reduced flavonols are regenerated. The transfer of electrons and protons to hydrogen peroxide are indicated above the dashed arrow. Types of enzymes that may catalyze these chemical reactions are presented in parentheses. It is also possible that the quinone reductase may convert the quinone back to a semiquinone. C. This diagram summarizes environmental or hormonal factors that increase levels of reactive oxygen species (ROS) and the role of flavonols as modulators of ROS to control developmental and physiological responses. Factors affecting ROS levels are indicated in colored boxes and the resulting changes in ROS concentration are depicted by increased size of boxes. Hormonal and environment increases in ROS levels are indicated by arrows and the ability of flavonols to reduce ROS accumulation is indicated by a blunt-end arrow. Observed developmental or physiological changes linked to the different ROS levels are indicated below the ROS changes. Abbreviation: ABA, abscisic acid.
