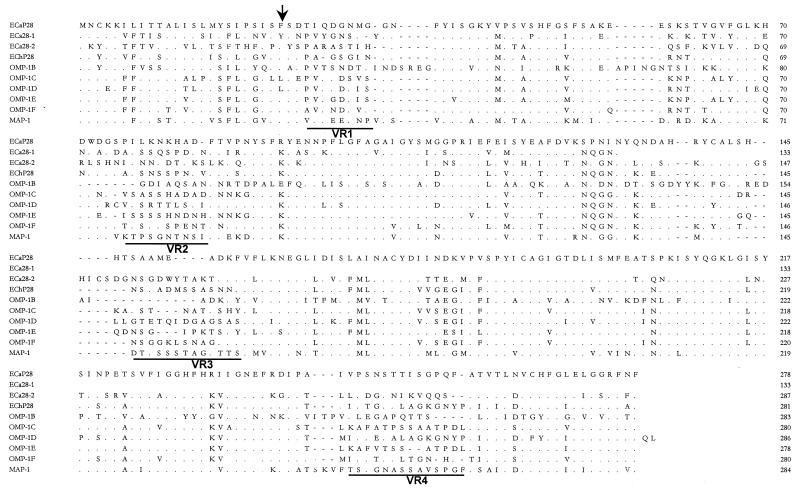
FIG. 3.
Alignment of E. canis P28 (ECaP28), E. canis 28-kDa protein 1 (ECa28-1) (complete) and 28-kDa protein 2 (ECa28-2) (partial), E. chaffeensis (ECh) OMP-1 family, and C. ruminantium MAP-1 amino acid sequences. The E. canis P28 amino acid sequence is presented as the consensus sequence. Amino acids not shown are identical to those of E. canis P28 and are represented by dots. Divergent amino acids are shown with the corresponding one-letter abbreviations. Gaps introduced for maximal alignment of the amino acid sequences are denoted with dashes. Variable regions are underlined and denoted VR1, VR2, VR3, and VR4. The arrow indicates the predicted signal peptidase cleavage site for the signal peptide.