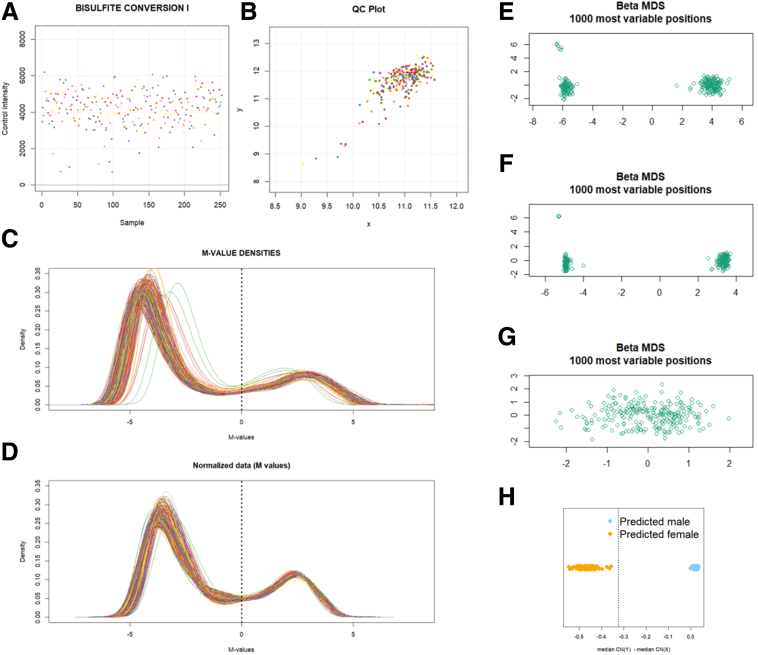
Figure 2.
ShinyMethyl output for quality control for TOPPIC methylation data. (A) Average negative control probe intensities. (B) Median intensity of M channel against median intensity of the u channel. (C, D) M-value intensities before and after functional normalization. (E–G) MDS during processing steps. (E) Raw data. (F) Following quantile normalization. (G) Following filtering of SNPs and sex chromosomes. (H) ShinyMethyl sex-prediction plot. No samples were mismatched for sex.