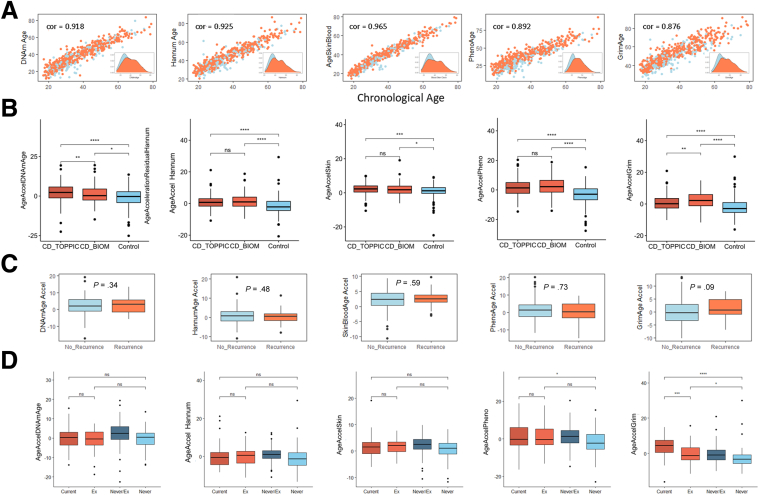
Figure 8.
Epigenetic age analysis using methods by (from right to left) Horvath (DNAmAge),40Hannum,41tissue specific (skin and blood clock),21phenoAge,42and GRIMage clocks.22 (A) Correlation plot of methylation age (y-axis) and biologic age (x-axis) using methods above, inset, density plot of methylation age). Cor, Pearsons R Correlation estimate. (B) Boxplots of age acceleration using methods above in patients with Crohn’s disease requiring surgery (CD_TOPPIC), newly diagnosed Crohn’s disease patients (CD_BIOM), and control subjects. (C) Boxplots of age acceleration in patients included in the TOPPIC trial who went on to develop recurrence or no recurrence following surgery. (C) Box plot for each methylation clock age acceleration and smoking status, current, exsmoking (recorded in the BIOM cohort), exsmoker/never smoker (grouped together as part of the TOPPIC cohort), and never smoked (recorded in the BIOM cohort). Ns = P > .05, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001 (Wilcox test).