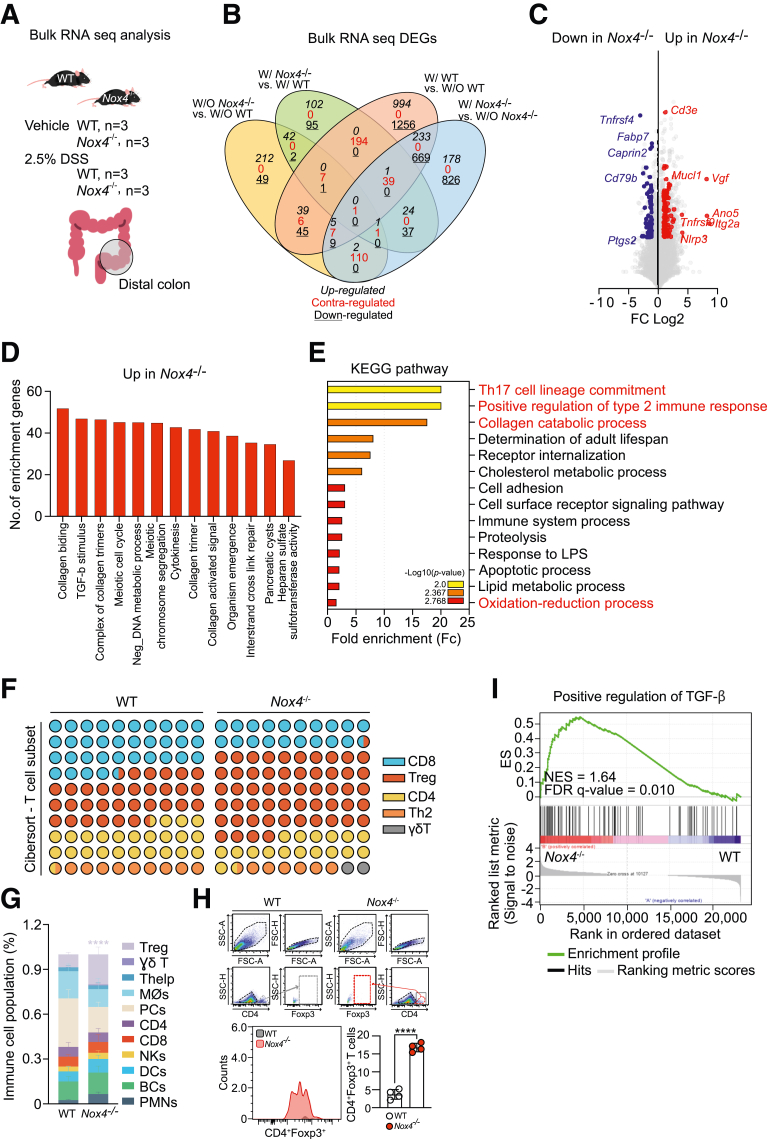
Figure 5.
Loss of Nox4 leads to collagen synthesis and Treg lineage commitment. (A) Scheme for bulk RNA-seq analysis of freshly isolated distal colon tissues comparing transcriptomes from the vehicle group in WT (n = 3) and Nox4-/- (n = 3) mice or the 2.5% DSS-treated colitis group in WT (n = 3) and Nox4-/- (n = 3) mice. (B) Venn diagram analysis of the 5124 DEGs comparing the numbers of up-regulated, contraregulated, and down-regulated within each control and experimental group. (C) Volcano plot identifying genes significantly down-regulated (blue) and up-regulated (red) with a more than 2-fold express level difference in Nox4-/- compared with WT samples. (D) GO biological process analysis of up-regulated DEGs in Nox4-/- mice. (E) KEGG pathway enrichment analysis of Nox4-/- DEGs. The top 14 pathways are identified, and the colors indicate statistical significance. (F) T-cell subpopulation evaluated by CIBERSORT based on WT and Nox4-/- DEGs. (G) CIBERSORT analysis of WT and Nox4-/- colon tissue. The relative proportions of 11 different immune cell types are deconvoluted from the RNA-seq data using CIBERSORT. (H) Flow cytometry analysis of freshly isolated WT or Nox4-/- distal colon tissue showing the relative number of live cells gated on CD4+Foxp3+ T cells (top, histogram) and the number of CD4+Foxp3+ T cells (bar graph, bottom). (I) Gene set enrichment analysis based on WT or Nox4-/- DEGs. Positive regulation of the TGF-β gene set is enriched with genes up-regulated in the Nox4-/- mouse colon. BC, B cell; DC, Dendritic cell; FDR, False discovery rate; FSC-A, Forward scatter-area; FSC-H, Forward scatter-height; ES, Enrichement score; LPS, lipopolysaccharide; NES, Normalized enrichment socre; NK, Natural killer; PC, Plasma cell; PMN, Polymorphonuclear leukocyte; SSC-A, Side scatter-area; SSC-H, Side scatter-height.